
miR-155 modulates fatty acid accumulation by targeting C/EBPβ in free fatty acid-induced steatosis in HepG2 cells
Introduction
Non-alcoholic fatty liver disease (NAFLD) is characterized by aberrant and non-alcoholic lipid droplet deposition in hepatocytes. The worldwide prevalence of NAFLD is now increasingly rising among the general population. About 25% of the world population, including children, was estimated to have suffered NAFLD in 2018 (1). A minority of individuals with non-alcoholic steatohepatitis (NASH), one subtype of NAFLD, are prone to advance to degenerative fibrosis, irreversible cirrhosis, and even hepatocellular carcinoma (2). NAFLD-related socioeconomic burdens and health-related quality of life (HR-QOL)-associated issues significantly affect patients’ outcomes. However, the pathogenesis of NAFLD is heterogeneous and still not well elucidated (3).
Patients with NAFLD display a high concentration of circulating free fatty acid (FFA) levels (4). In addition, a high concentration of circulating FFA levels can cause a sequence of pathological processes varying from insulin resistance, diabetes, and ultimately even to hepatic steatosis (5,6). Therefore, cellular FFA overloading is commonly applied to mimic hepatic steatosis with in vitro experimental models (7,8). In this study, we utilized FFA-loaded HepG2 cells to construct an in vitro model of NAFLD and then investigated the potential underlying molecular mechanisms of NAFLD.
MicroRNAs (miRNAs, miRs) are a class of small endogenous noncoding RNAs that contain approximately 19–22 nucleotides, which negatively regulate their target genes by directly binding to the 3’untranslated region (3’UTR) of related mRNAs via complementary base pairing principle and extensively participating in various pathophysiological events in the liver (9-11). It has been reported that miR-122 is the most abundant miRNA in the normal human liver, and circulating miR-122, miR-192 and miR-375 were upregulated in benign steatohepatitis and even more dramatically increased in NASH (11). Additionally, silencing of miR-34a was found to restore the expression of SIRT1 and PPARα, leading to AMPK and HMGCR activation, finally ameliorating hepatic steatosis (12). In human NASH, palmitic acids-treated HepG2 cells, or in high fat diet (HFD)-fed mice, downregulation of miR-451 could inhibit fatty acid-induced effects through the regulation of AMPK/AKT pathway (13). These discoveries suggest that miRNAs play a pivotal role in NAFLD-associated pathogenesis.
miR-155 has been found to be increased in liver tissues from HFD-fed mice (14), indicating that differentially expressed miR-155 may be involved in the regulation of lipid metabolism. Notably, recent investigations revealed that miR-155 is a multifunctional miRNA, and plays pivotal roles in a variety of physiological and pathological biology processes, including haematopoiesis, inflammation, hematological and solid malignancies, and cardiovascular diseases (15-18). In addition, another study demonstrated that miR-155/LXRα axis played a protective role in NAFLD mice (19). Conditionally liver-specific overexpression of miR-155 ameliorated HFD-induced fatty liver in mice by targeting carboxylesterase 3/triacylglycerol hydrolase (Ces3/TGH) (20). Due to the relevance between miR155 and NAFLD, we set out to perform an in-depth analysis of miR-155 in HepG2 cells treated with FFA with the aim of finding a new target of miR-155 responsible for the pathogenesis of NAFLD.
We were subsequently able to demonstrate that miR-155 was upregulated in FFA-exposed HepG2 cells in vitro. Interestingly, in the absence of FFA exposure, inhibition of miR-155 was capable of promoting fatty acid accumulation in HepG2 cells; meanwhile, in the presence of FFA exposure, overexpression of miR-155 attenuated this accumulation in HepG2 cells. Further investigation revealed that CCAAT/enhancer binding protein β (C/EBPβ) was a target gene of miR-155, suggesting that targeting miR-155/C/EBPβ might represent an attractive strategy for preventing NAFLD.
Methods
HepG2 Cells culture, transfection and FFA treatment
Human liver hepatocellular carcinoma (HepG2) cells were purchased from the cell bank of The Chinese Academy of Sciences (Shanghai, China). Cells were maintained in high glucose-DMEM (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1% penicillin/streptomycin. Cells were cultured at 37°C in an incubator supplied with 5% CO2.
For cell transfection, HepG2 cells were starved by 1% FBS-containing medium for 6 hrs, and thereafter transfected with miR-155 mimics (50 nM, RiboBio, Guangzhou, China), miR-155 inhibitor (100 nM, RiboBio), or relative negative controls for 48 hrs by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). C/EBPβ siRNAs (75 nM, RiboBio) and non-target control siRNAs were transfected in HepG2 cells or transfected in HepG2 cells in combination with miR-155 inhibitor for 48 hrs.
For FFA treatment, 24 hrs after transfection, HepG2 cells were exposed to a mixture consisting of 1 mM oleate and palmitate at a molar ratio of 2:1 (Sigma-Aldrich, St. Louis, MO, USA) which was prepared in 1% bovine serum albumin (BSA)-containing medium (KeyGEN, Nanjing, China) for 24 hrs as previously described (14).
Nile red staining
HepG2 cells were seeded in 24-well plates. After exposure with 1 mM of FFA for 24 hrs, HepG2 cells were fixed and then stained with lipophilic fluorescent Nile red dye (0.1 µM) for 15 mins at room temperature. Cell nuclei were stained with Hoescht-33342. After this, fluorescence images were acquired on a fluorescence microscope (Leica, Wetzlar, Germany) under 100× magnification.
Flow cytofluorometry for determination of intracellular lipid droplet content
Forty-eight hours after treatment, HepG2 cells were collected. HepG2 cells were centrifuged at 166 g for 5 min, resuspended in 1 mL PBS and incubated with 0.75 µg/mL. Nile red dye was used for 15 min at room temperature. Intracellular lipid droplet content was analyzed by flow cytometer (Beckman, Miami, FL, USA) on the FL3 emission channel through a 585±21 nm band pass filter, following excitation with an argon ion laser source at 488 nm. Fluorescence intensity was quantified to calculate the percentage of events above the median value of fluorescence. The data was analyzed by FlowJo software (Treestar Inc, Ashland, OR, USA) and represented as the fold change (FC) comparing treatment groups to control groups.
RNA extraction and quantitative real time-polymerase chain reaction (qRT-PCR)
HepG2 cells were seeded at a density of 1×106 per well in 12-well plates. After treatment, cells were harvested. Total RNA was isolated from HepG2 cells using TRIzol reagent (TaKaRa, Kusatsu, Japan). For the analysis of C/EBPβ mRNA, complementary DNA (cDNA) was reverse transcribed from total RNA by iScriptTM cDNA Synthesis Kit (Bio-Rad, Richmond, CA, USA). Subsequent amplification and examination were performed with SYBR Green (Takara) based on the ABI 7900HT Fast Real-Time PCR System (Bio-Rad). The primers of C/EBPβ and 18s are listed in Table 1. For the quantificative analysis of miR-155, the Bulge-LoopTM miRNA qPCR Primer Set (RiboBio) was applied to determine the expression of miR-155 with SYBR Green. qRT-PCRs were performed based on ABI 7900HT Fast Real-Time PCR System. The relative mRNA and miRNA expression levels were determined by the 2-ΔΔCt method. Housekeeping genes 18s and U6 were respectively used as the internal control for C/EBPβ mRNA and miR-155 expression analysis.
Table 1
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| has-CEBPB | CTTCAGCCCGTACCTGGAG | GGAGAGGAAGTCGTGGTGC |
| has-18S rRNA | CTTTCGAGGCCCTGTAATTG | CCTCCAATGGATCCTCGTTA |
Western blotting
HepG2 cells were seeded at a density of 1×106 per well in 6-well plates. After treatment, cells were harvested and then lysed on ice in RIPA lysis buffer (KeyGEN) containing 1% phenylmethanesulfonyl fluoride (PMSF) (KeyGEN). Equivalent amounts of total protein were subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride (PVDF) membranes (Millipore), and immunoblotted with primary monoclonal antibodies for C/EBPβ (1:1,000 dilution, ABclonal, Cambridge, MA, USA) at 4 °C overnight. The membranes were then incubated with corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000 dilution, Bioworld) at room temperature for 2 hrs. Band intensity was visualized by enhanced chemiluminescence (ECL) system (Tanon, Shanghai, China) using Image J software (NIH, Bethesda, MD, USA). β-actin (1:10,000 dilution, ABclonal) was used as a loading control.
Statistical analysis
All statistical analyses were performed by SPSS version 22.0 software (IBM, Armonk, NY, USA). Unpaired, two-tailed Student’s t-test was used for two-group comparation. One-way ANOVA was used for comparing four groups followed by Bonferroni correction. A P value <0.05 was defined to be a statistically significant value. The data are presented as mean ± standard error of mean (SEM).
Results
MiR-155 is increased in FFA-exposed HepG2 cells
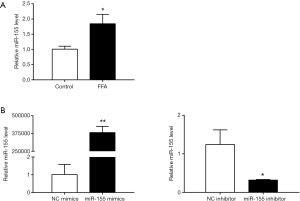
HepG2 cells were exposed to free fatty acid (FFA) (oleate: palmitate at the molar ratio of 2:1) for 24 hrs to induce hepatic steatosis with in vitro mimicking NAFLD. The miR-155 level was significantly upregulated in FFA-exposed HepG2 cells (Figure 1A), indicating that miR-155 was dysregulated in FFA-exposed HepG2 cells and appeared to be a contributor to NAFLD pathogenesis.
MiR-155 affects intracellular lipid droplet content in HepG2 cells
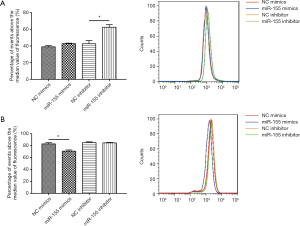
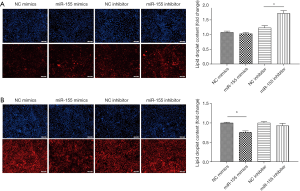
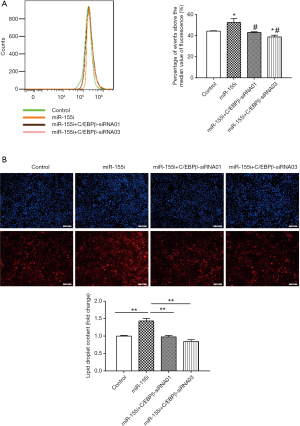
To determine the functional role of miR-155 on intracellular lipid droplet content alteration in vitro, HepG2 cells were transfected with miR-155 mimics, inhibitor, or negative controls. As shown in Figure 1B, miR-155 mimics increased, whereas inhibitor decreased, the miR-155 expression level. The gain-of-function and loss-of-function experiments were performed in HepG2 cells in the presence or in the absence of FFA exposure. Generally, in the absence of FFA exposure, flow cytofluorometry showed that knockdown of miR-155 significantly promoted lipid droplet content in HepG2 cells, whereas overexpression of miR-155 could not attenuate lipid droplet content (Figure 2A). In the presence of FFA exposure, flow cytofluorometry showed that overexpression of miR-155 reduced lipid droplet accumulation, but knockdown of miR-155 could not further promote lipid droplet accumulation (Figure 2B). In summary, these results indicated that overexpression miR-155 negatively regulated lipid droplet accumulation when the HepG2 cells were overloaded with FFA; in contrast, inhibition of miR-155 could positively promote lipid droplet content when the HepG2 cells were at basal level of lipid accumulation. These results are consistent with those obtained by fluorescence microscope imaging (Figure 3). In conclusion, these findings confirm the role of miR-155 in intracellular lipid droplet content alteration in vitro.
C/EBPβ is a target gene of miR-155 involved in lipid droplet accumulation
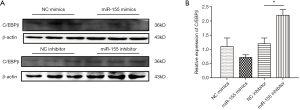
To identify the target gene of miR-155, we firstly browsed online the miRNA target prediction database. Based on both the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/) and the PicTar database (https://pictar.mdc-berlin.de/), we noticed that C/EBPβ was a predicted target gene of hsa-miR-155. Knowing that C/EBPβ is well documented for its roles in modulating hepatocytic proliferation and survival, and its emerging role in the regulation of hepatic lipogenesis, we continued to investigate if C/EBPβ was a target gene of miR-155 involved in lipid droplet accumulation. miR-155 mimics had the tendency to decrease C/EBPβ while miR-155 inhibitor could elevate C/EBPβ (Figure 4), suggesting C/EBPβ might be a target gene of miR-155.
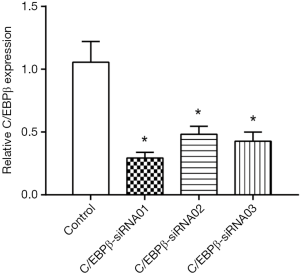
SiRNAs targeting C/EBPβ were used in a rescue assay to further evaluate whether the functional roles of miR-155 in the lipid droplet accumulation of HepG2 cells were mediated by C/EBPβ. We firstly validated the effects of C/EBPβ siRNAs by qRT-PCR (Figure 5). C/EBPβ siRNA01 and C/EBPβ siRNA03 were used in the other studies. Silencing C/EBPβ reversed the increased lipid droplet content induced by miR-155 inhibitor in HepG2 cells (Figure 6). Thus, C/EBPβ is a target gene of miR-155, which is responsible for the effects of miR-155 involved in lipid droplet accumulation.
Discussion
NAFLD is currently the most common liver disease in the world, but the pathogenesis of NAFLD is still unclear. As stated by the well-known double-hit theory, the first hit is the aberrant accumulation of triglycerides in hepatocytes, and the second hit is the oxidative stress and inflammatory mediators advancing hepatosteatosis to steatohepatitis, fibrosis, and even cirrhosis. However, recent findings have supported the multiple-hit hypothesis as being more powerful in explaining NAFLD pathogenesis. Multiple pathogenic factors, such as lipotoxicity, adipose tissue dysfunction, insulin resistance, oxidative stress, mitochondrial dysfunction, endoplasmic reticulum stress, altered innate immune regulation, and cytokine secretion, contribute to liver injury (21). Hallmarked by hepatic steatosis, which is a consequence of lipid acquisition surpassing lipid disposal, NAFLD is characterized by lipid homeostasis dysregulation. Little is known about its underlying molecular mechanisms. To date, there is no effective treatment for NAFLD. On account of this, there is an urgent need for these molecular mechanisms to be comprehensively elucidated, and for the corresponding NAFLD therapeutics to be innovated.
Currently, 38589 human miRNAs have been annotated (miRBase: http://www.mirbase.org/index.shtml, release 22.1, October 2018), and it is estimated that up to 60% of all human protein-coding genes might be direct targets of currently known miRNAs (22). The dysregulation of hepatic microRNAs has been commonly observed in NAFLD, and numerous dysregulated miRNAs are reported to be responsible for the development of NAFLD [e.g., miR-21 targets HMGCR to control triglyceride and cholesterol metabolism in NAFLD (23), miR-296 regulates FFA-induced lipoapoptosis by targeting PUMA in vitro (24), miR375 controls adipokines and inhibited inflammatory cytokines by directly targeting AdipoR2 in palmitic acid-induced HepG2 cells (25), and miR-149 targets FGF-21 to regulate NAFLD development] (26).
miR-155 has been reported to protect mice from the development of non-alcoholic hepatosteatosis by targeting LXRα (19). Circulating miR-155 decreases remarkably in NAFLD patients and could serve as a biomarker for NAFLD (27), indicating that miR-155 is closely related to NAFLD pathogenesis. However, the function of miR-155 in FFA-induced hepatic steatosis model of NAFLD in vitro has not yet been reported. Nile red is commonly utilized as an excellent stain for the investigation of physiological lipid droplet formation and pathological lipid overloading in cells (28,29). Thus, by using flow cytofluorometric analysis and fluorescence microscopy imaging, we investigated the effects of miR-155 in vitro based on FFA-exposed hepatic steatosis model. Our results demonstrated that overexpression of miR-155 is capable of attenuating lipid accumulation in HepG2 cells in the presence of FFA exposure, while inhibition of miR-155 is capable of promoting lipid accumulation in the absence of FFA exposure.
C/EBPβ is a member of the liver-enriched transcription factors C/EBPs family. The basic leucine zipper (bZIP) domain of C/EBPβ is highly conserved and primarily contributes to dimerization and DNA binding. Two members of the C/EBPs family, C/EBPα and C/EBPβ, are central governors of the integrative metabolic biological processes in the liver, such as gluconeogenic genes. Besides this, binding motifs of C/EBPs exist in many lipogenic gene promoters (30), suggesting that C/EBPβ might be a crucial mediator for lipid accumulation in the liver. C/EBPβ participates in forming LXRα-C/EBPβ complex which binds to the promoter of SREBP-1c. And the complex activates SREBP-1c transcription which ultimately activates genes encoding enzymes required for fat synthesis and lipid metabolism regulation in the liver (31). In palmitate-exposed HepG2 cells, knockdown of C/EBPβ alleviates lipid deposition and boosts lipolysis (32), indicating that C/EBPβ might be a crucial factor involved in lipid metabolism in the liver. Tumor necrosis factor α (TNFα) induces up-regulation of miR-155 and can significantly inhibit adipogenesis via targeting C/EBPβ (33). Herein, we report that the lipid droplet accumulation-inducing effect of miR-155 inhibitor can be reversed by interfering C/EBPβ in HepG2 cells. Thus, C/EBPβ is a target gene of miR-155 responsible for lipid accumulation in HepG2 cells.
Since the transcription factor C/EBPβ has been extensively reported to transcriptionally regulate key genes that control metabolic processes including lipogenesis and gluconeogenesis, finding molecules that are transcriptionally regulated by C/EBPβ or molecules that crosstalk with C/EBPβ and constructing a regulatory network of lipid accumulation would contribute to further in-depth mechanism study. Peroxisome proliferator-activated receptors (PPARs) are known as the master regulators of lipid metabolism, glucose metabolism, and energy homeostasis in the liver. The fact that CCAAT/enhancer-binding protein (C/EBP)-binding motifs exist adjacent to most PPARγ-binding sites highlights that PPARγ and C/EBPβ are likely part of a complex transcriptional network that regulates gene expression in a spatially coordinated manner (34). Fibroblast growth factor (FGF21), acting as an endocrine hormone, is secreted by the liver in response to peroxisome proliferator-activated receptor-α (PPARα) activation, exerting its metabolic effects mostly in the liver. PPARα/FGF21 signaling pathway has been recently reported to be a hepatic-specific mechanism in the pathogenesis of NAFLD (35). The intricate regulatory network between miR-155/C/EBPβ axis and some other closely related molecules including PPARγ or PPARα/FGF21 pathway needs to be precisely clarified.
Reliable and definite animal models are crucial for further investigation because in-vivo NAFLD models possess features resembling those of both the histopathology and pathophysiology of the stages and progression of NAFLD. Investigation into the function of miR-155/C/EBPβ in mouse NAFLD model is also urgently required. Distinct from the extensively used classic dietary models which mainly included high-fat diet, methionine-choline deficient diet (MCDD) and high-fructose (HF) diet, the western diet (WD) combined with carbon tetrachloride (CCl4) injections (WD/CCl4) has been recently noted for its inherent advantages in NAFLD pathogenesis (36). Thus, constructing a WD/CCL4 dietary model should be taken into account in the future research of relevant mechanisms, along with the ultimate goal of drug development.
In summary, our study showed that miR-155 was elevated in the FFA-induced hepatic steatosis model of NAFLD in vitro. Additionally, MiR-155 regulated fatty acid accumulation by targeting C/EBPβ. Hence, pharmacological intervention of miR-155 that decreases hepatic lipid contents represents a potential therapeutic strategy for NAFLD.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.02.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments and methods are performed strictly in accordance with the Declaration of Helsinki and the relevant guidelines of Wenzhou Medical University. The institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Younossi Z, Tacke F, Arrese M, et al. Global Perspectives on Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis. Hepatology 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Febbraio MA, Reibe S, Shalapour S, et al. Preclinical Models for Studying NASH-Driven HCC: How Useful Are They? Cell Metab 2019;29:18-26. [Crossref] [PubMed]
- Younossi ZM, Henry L. Economic and Quality-of-Life Implications of Non-Alcoholic Fatty Liver Disease. Pharmacoeconomics 2015;33:1245-53. [Crossref] [PubMed]
- Zhang J, Zhao Y, Xu C, et al. Association between serum free fatty acid levels and nonalcoholic fatty liver disease: a cross-sectional study. Sci Rep 2014;4:5832. [Crossref] [PubMed]
- Natarajan SK, Ingham SA, Mohr AM, et al. Saturated free fatty acids induce cholangiocyte lipoapoptosis. Hepatology 2014;60:1942-56. [Crossref] [PubMed]
- Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis 2008;28:360-9. [Crossref] [PubMed]
- Castro RE, Ferreira DM, Afonso MB, et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol 2013;58:119-25. [Crossref] [PubMed]
- Chen Y, Rachek L, Shadel G, et al. 515 - Oxidized Mitochondrial DNA Induces Steatohepatitis through CGAS Signaling Pathway. Gastroenterology 2018;154:S-1096 [Crossref]
- Valdmanis PN, Kim HK, Chu K, et al. miR-122 removal in the liver activates imprinted microRNAs and enables more effective microRNA-mediated gene repression. Nat Commun 2018;9:5321. [Crossref] [PubMed]
- Xu L, Li Y, Yin L, et al. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics 2018;8:5593-609. [Crossref] [PubMed]
- Liu XL, Cao HX, Wang BC, et al. miR-192-5p regulates lipid synthesis in non-alcoholic fatty liver disease through SCD-1. World J Gastroenterol 2017;23:8140-51. [Crossref] [PubMed]
- Ding J, Li M, Wan X, et al. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci Rep 2015;5:13729. [Crossref] [PubMed]
- Hur W, Lee JH, Kim SW, et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int J Biochem Cell Biol 2015;64:265-76. [Crossref] [PubMed]
- Xiao J, Bei Y, Liu J, et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J Cell Mol Med 2016;20:204-16. [Crossref] [PubMed]
- Georgantas RW 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A 2007;104:2750-5. [Crossref] [PubMed]
- Zitzer NC, Snyder K, Meng X, et al. MicroRNA-155 Modulates Acute Graft-versus-Host Disease by Impacting T Cell Expansion, Migration, and Effector Function. J Immunol 2018;200:4170-9. [Crossref] [PubMed]
- Elmesmari A, Fraser AR, Wood C, et al. MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in Rheumatoid Arthritis. Rheumatology (Oxford) 2016;55:2056-65. [Crossref] [PubMed]
- Jurkovicova D, Magyerkova M, Kulcsar L, et al. miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma 2014;61:241-51. [Crossref] [PubMed]
- Miller AM, Gilchrist DS, Nijjar J, et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One 2013;8:e72324 [Crossref] [PubMed]
- Lin X, Jia J, Du T, et al. Overexpression of miR-155 in the liver of transgenic mice alters the expression profiling of hepatic genes associated with lipid metabolism. PLoS One 2015;10:e0118417 [Crossref] [PubMed]
- Wang T, Pan W, Hu J, et al. Circular RNAs in Metabolic Diseases. Adv Exp Med Biol 2018;1087:275-85. [Crossref] [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Sun C, Huang F, Liu X, et al. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med 2015;35:847-53. [Crossref] [PubMed]
- Cazanave SC, Mott JL, Elmi NA, et al. A role for miR-296 in the regulation of lipoapoptosis by targeting PUMA. J Lipid Res 2011;52:1517-25. [Crossref] [PubMed]
- Lei L, Zhou C, Yang X, et al. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin Exp Pharmacol Physiol 2018;45:819-31. [Crossref] [PubMed]
- Xiao J, Lv D, Zhao Y, et al. miR-149 controls non-alcoholic fatty liver by targeting FGF-21. J Cell Mol Med 2016;20:1603-8. [Crossref] [PubMed]
- Wang L, Zhang N, Wang Z, et al. Decreased MiR-155 Level in the Peripheral Blood of Non-Alcoholic Fatty Liver Disease Patients may Serve as a Biomarker and may Influence LXR Activity. Cell Physiol Biochem 2016;39:2239-48. [Crossref] [PubMed]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 1985;100:965-73. [Crossref] [PubMed]
- Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res 1985;26:781-9. [PubMed]
- Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 2004;56:291-330. [Crossref] [PubMed]
- Tian J, Goldstein JL, Brown MS. Insulin induction of SREBP-1c in rodent liver requires LXRα-C/EBPβ complex. Proc Natl Acad Sci U S A 2016;113:8182-7. [Crossref] [PubMed]
- Zhao NQ, Li XY, Wang L, et al. Palmitate induces fat accumulation by activating C/EBPbeta-mediated G0S2 expression in HepG2 cells. World J Gastroenterol 2017;23:7705-15. [Crossref] [PubMed]
- Liu S, Yang Y, Wu J. TNFalpha-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem Biophys Res Commun 2011;414:618-24. [Crossref] [PubMed]
- Lefterova MI, Zhang Y, Steger DJ, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 2008;22:2941-52. [Crossref] [PubMed]
- Piccinin E, Moschetta A. Hepatic-specific PPARalpha-FGF21 action in NAFLD. Gut 2016;65:1075-6. [Crossref] [PubMed]
- Castro RE, Diehl AM. Towards a definite mouse model of NAFLD. J Hepatol 2018;69:272-4. [Crossref] [PubMed]
Cite this article as: Hu J, Chen X. miR-155 modulates fatty acid accumulation by targeting C/EBPβ in free fatty acid-induced steatosis in HepG2 cells. Non-coding RNA Investig 2019;3:10.


