
Long noncoding RNA SYISL: the crucial interaction with EZH2 in skeletal muscle differentiation and disorders
Epigenetics is a branch of genetics that studies the heritable phenotype changes influencing gene expression, cellular functions and fate, which do not occur through altered DNA sequence of the genes. The investigation of the underlying mechanisms of epigenetics and the deep understanding of its functions in the biological process are milestones for biology research.
The main systems that initiate and sustain epigenetics changes include DNA methylation, histone modification and non-coding RNA (ncRNA)-associated gene silencing. In the last few years, the studies regarding the function of ncRNAs have attracted remarkable interest (1,2), however, despite significant progresses, the identification of the roles of ncRNAs is still a great challenge for the scientific community.
The ncRNAs comprise a rich variety of regulatory and functional non-coding regions, including transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, snRNAs, and the long ncRNAs (lncRNAs).
The lncRNAs are a heterogeneous class of ncRNAs, poorly characterized, including thousands of different units. With the development of innovative technologies dedicated to exploring the transcriptome, considerable improvement in the identification and characterization of lncRNAs has been made (3,4). New functions for lncRNAs are emerging, suggesting a key role of lncRNAs in the epigenetic regulation of several biological process, including cell proliferation and differentiation (5,6).
Using high-throughput RNA sequencing (RNA-seq) approaches, the expression level of lncRNAs has been evaluated and it was found to be more tissue- and cell type-specific compared to protein-coding genes (7,8). Several lncRNAs have been linked to human diseases, such as coronary artery diseases, autoimmune diseases, neurological disorders and various cancers (6,9).
Jin et al. focused their study on the investigation of lncRNAs function in the myogenesis process and in the regulation of muscle mass, since this topic is still unclear (10). The myogenesis is a complex process controlled by several muscle-specific transcription factors and epigenetics regulators (Figure 1A). Recent studies suggest a crucial role of lncRNAs in skeletal muscle cell differentiation and muscle development (11,12). A lncRNA region was recently found to be involved in promoting myogenesis and enhancing muscle mass in vivo (13).
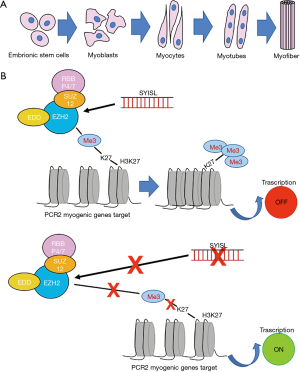
Following this path, Jin et al., through the characterization of expression profiles of lncRNAs during C2C12 myoblast differentiation, were able to identify an intronic lncRNA named “SYISL” (SYNPO2 intron sense-overlapping lncRNA), involved in the repression of muscle development (10). In order to identify target genes regulated by SYISL, knockdown and genome-wide expression approaches have been used and correlation between SYISL region with muscle differentiation and disease associated pathways have been recognized. After SYISL knockdown, several genes were found to be significantly up-regulated, such as MyoG, Myh1, Myh2, Myh4, Myh7, Tnni1, and Mybpc2.
Moreover, functional gain and loss analysis were used to investigate the effects of SYISL on myoblast proliferation and differentiation in C2C12 cells. Interestingly, SYISL acted as a promoter of myoblast proliferation inducing myoblast fusion into one myotube. However, at the same time, it was noticed that SYISL was involved in an impaired cell migration accompanied by a reduced cell differentiation. The SYISL knockout (KO) significantly increased the myogenic differentiation of C2C12 cells, as demonstrated by an overexpression of mRNA and protein levels of the myogenic marker genes such as MyoG and MyHC. On the other hand, the SYISL overexpression significantly decreased the expression of these myogenic genes, with repression of cell migration.
Surprisingly, analyzing SYISL-KO mice compared to wild-type (WT) mice, a statistically significant intensification of muscle density and mass was identified, with an increased number of smaller muscle fibers. As suggested by the authors, these conditions might be due to the elimination of myogenic gene silencing mediated by SYISL. However, the mice SYISL-KO model revealed a stimulation of muscle regeneration, correlated to an earlier myogenic differentiation potential.
Based on these evidences, lncRNA, and, in this specific case SYISL, can be considered fundamental for the regulation of the muscle differentiation, pointing out the need to better understand their mechanisms of action.
As revealed by recent studies, the myogenic process is associated with global changing in chromatin, especially correlated to polycomb repressive complex 2 (PRC2) (14,15). PRC2 is a protein-complex involved in epigenetic gene silencing, required in many processes including cell proliferation, differentiation, stem cell maintenance, embryonic development and also myogenic differentiation (16,17). An extremely interesting finding reported by Jin et al. is the direct interaction between SYISL and PRC2 with the consequent inhibition of myogenic differentiation (10) (Figure 1B). PRC2 is composed by enhancer of zeste homolog 1 and 2 (EZH1/2), suppressor of zeste 12 (SUZ12), the embryonic ectoderm development (EED) and retinoblastoma-binding protein 4/7 (RBBP4/7) (18). EZH2 is the catalytic subunits of this complex, which is able to repress gene transcription through the tri-methylation of histone H3 lysine (H3K27me3) (19). EZH2 exerts a critical role in the regulation of skeletal muscle development, inhibiting the transcription of muscle-specific genes and cell-cycle genes, preventing the premature differentiation (15,20).
Jin et al. confirmed that the overexpression of EZH2 gene stimulates the cell proliferation and induces a decrease of myogenic differentiation (10). By using several assays, the interaction between SYISL and EZH2 was evaluated, observing the role of SYISL in the recruitment of EZH2 to the promoters of its myogenic target genes (Figure 1B). SYISL influences the binding capacity of EZH2 at MyoG, Myh4, and MCK gene promoters, increasing the enrichment of EZH2 at these sites. Indeed, with SYISL-KO, a removal of EZH2 from its targets has been shown. Moreover, the full-length SYISL was found to be strictly necessary to interact with EZH2 and, therefore, to repress myogenic differentiation, suggesting the importance of the higher-order structures of SYISL to fulfil its function. Furthermore, the stimulation of myoblast proliferation correlated to SYISL was supported by the inhibition of p21 gene expression mediated by recruiting EZH2. According to these results, others lncRNAs were found capable to interact with PRC2 and to recruit it to the specific regulatory regions, inducing epigenetics silencing (21,22).
The indispensable role of lncRNAs and, in particular, of SYISL in the EZH2 activity is of extreme interest for the study of the myogenic process and, therefore, for the diseases correlated with it. Rhabdomyosarcoma (RMS), a pediatric tumor arisen from muscle precursor cells, is a relevant example of an aggressive myogenic disorder. In RMS, the muscle cells show an abnormal proliferation and altered differentiation program that leads to an incomplete myogenesis (23). Several studies have already shown the crucial role of EZH2 in the impaired differentiation state of muscle cells in RMS (15,24). The EZH2-knockdown in RMS cells line promoted reactivation of myogenic genes with a restore of myocyte phenotype (25).
As future prospective, the investigation of the role of SYISL in myogenic diseases could be particularly relevant, such as RMS, analyzing its expression and interactions in pathological conditions. Among other things, it would be interesting to understand if SYISL is correlated to the altered muscle development associated with EZH2 in RMS, especially because, SYISL, during myogenesis, regulates the expression of some myogenic genes, particularly myocyte fusion-associated genes, not related to EZH2. Therefore, this suggests that SYISL could be acting through other mechanisms, which need to be elucidated.
These fascinating outcomes highlight the importance of lncRNAs in the epigenetic regulation of the myogenic development and cell proliferation. In this context, SYISL can be considered a relevant lncRNAs involved in the control of myoblast differentiation mediated by PRC2 and, in particular, EZH2. After analyzing in depth, through various and innovative approaches, the localization, interactions and the effects of SYLSL on myogenesis, Jin et al. proposed an intriguing explanation of its function. In any case, further analysis should be conducted, especially to clarify how SYISL works on muscle mass and density and to investigate the other mechanisms involved, which are yet unknown. Nevertheless, these emerging finding pave the way for new insights in the identification of the role of lncRNAs in biological processes.
Acknowledgments
Funding: V Bordoni is financially supported by the PhD School in Life Sciences and Biotechnologies at the University of Sassari (P.O.R. F.S.E. 2014-2020).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Dr. Rongrong Gao, Section Editor (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.01.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Guo X, Gao L, Wang Y, et al. Advances in long noncoding RNAs: identification, structure prediction and function annotation. Brief Funct Genomics 2016;15:38-46. [Crossref] [PubMed]
- Hu X, Sood AK, Dang CV, et al. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev 2018;48:8-15. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Li D, Yang MQ. Correction to: Identification and characterization of conserved lncRNAs in human and rat brain. BMC Bioinformatics 2018;19:181. [Crossref] [PubMed]
- Li J, Tian H, Yang J, et al. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol 2016;35:459-70. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci 2013;14:18790-808. [Crossref] [PubMed]
- Jin JJ, Lv W, Xia P, et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc Natl Acad Sci U S A 2018;115:E9802-11. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet Muscle 2014;4:8. [Crossref] [PubMed]
- Zhu M, Liu J, Xiao J, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun 2017;8:14718. [Crossref] [PubMed]
- Adhikari A, Davie J. JARID2 and the PRC2 complex regulate skeletal muscle differentiation through regulation of canonical Wnt signaling. Epigenetics Chromatin 2018;11:46. [Crossref] [PubMed]
- Marchesi I, Giordano A, Bagella L. Roles of enhancer of zeste homolog 2: from skeletal muscle differentiation to rhabdomyosarcoma carcinogenesis. Cell Cycle 2014;13:516-27. [Crossref] [PubMed]
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development 2013;140:2525-34. [Crossref] [PubMed]
- Sanna L, Marchesi I, Melone MA, et al. The role of enhancer of zeste homolog 2: From viral epigenetics to the carcinogenesis of hepatocellular carcinoma. J Cell Physiol 2018;233:6508-17. [Crossref] [PubMed]
- Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Molecular Cell 2008;32:503-18. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Caretti G, Di Padova M, Micales B, et al. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004;18:2627-38. [Crossref] [PubMed]
- Negishi M, Wongpalee SP, Sarkar S, et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One 2014;9:e95216 [Crossref] [PubMed]
- Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015;21:2007-22. [Crossref] [PubMed]
- Merlino G, Helman LJ. Rhabdomyosarcoma--working out the pathways. Oncogene. 1999;18:5340-8. [Crossref] [PubMed]
- Marchesi I, Sanna L, Fais M, et al. 12-O-tetradecanoylphorbol-13-acetate and EZH2 inhibition: A novel approach for promoting myogenic differentiation in embryonal rhabdomyosarcoma cells. J Cell Physiol 2018;233:2360-5. [Crossref] [PubMed]
- Marchesi I, Fiorentino FP, Rizzolio F, et al. The ablation of EZH2 uncovers its crucial role in rhabdomyosarcoma formation. Cell Cycle 2012;11:3828-36. [Crossref] [PubMed]
Cite this article as: Bordoni V, Bagella L. Long noncoding RNA SYISL: the crucial interaction with EZH2 in skeletal muscle differentiation and disorders. Non-coding RNA Investig 2019;3:7.


