
Senescence lncRNAs govern cell surface components: lncRNA-OIS1 transcriptionally elevates DPP4
With advancing age, senescent cells accumulate in tissues and organs, accelerating aging and age-related disease (e.g., diabetes, neurodegeneration, and cancers) (1). Cellular senescence is triggered by sublethal stresses including telomere shortening (replicative senescence), damage to DNA or other molecules (premature senescence), and oncogenic activation (oncogene-induced senescence, OIS) (2-5). Regardless of the trigger, all forms of senescence share common features like cell cycle arrest via p53 (TP53)/p21 (CDKN1A) and p16 (CDKN2A)/RB pathways, increased senescence-associated β-galactosidase (SA-β-Gal) activity, and the onset of a senescence-associated secretory phenotype (SASP) (6,7). OIS is tightly linked to tumor suppression, as it is elicited by unscheduled expression of oncogenic proteins such as HRAS, E2F1, RAF, BRAF, and MOS (8-11). The oncogenic protein HRASG12V (bearing a mutation of G to V at amino acid position 12 in HRAS) triggers senescence and is commonly used as a model to trigger OIS (12).
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that generally lack protein-coding potential. RNA sequencing (RNA-seq) analysis has identified a large number of lncRNAs expressed in various cells and developmental conditions (13-15). Although lncRNAs do not encode proteins, they are potent regulators of gene expression both at the transcriptional and post-transcriptional levels. They control transcription by modifying chromatin structure and recruiting transcriptional activators or repressors (16,17). For instance, the lncRNA HOTAIR (HOX transcript antisense RNA) is transcribed from the HOXC locus and mediates gene silencing of the HOXD locus by binding and recruiting the polycomb repressive complex 2 (PRC2) (18). Unlike HOTAIR, lncRNA Evx1as enhances EVX1 transcription by binding to Evx1as/EVX1 enhancer site and looping the chromatin between the promoter and enhancer. This conformation facilitates the assembly of machineries required for efficient EVX1 transcription (19). Post-transcriptionally, lncRNAs regulate gene expression in many different ways. They can form scaffolds enabling protein assembly of complexes and act as decoys to regulate the availability of microRNAs and RNA-binding proteins (RBPs) to mRNAs (20,21). They can also influence the formation of ribonucleoprotein complexes encompassing mRNAs and RBPs (22,23); for example, 7SL binds the 3’ untranslated region (3’UTR) of TP53 mRNA and suppresses expression of the tumor suppressor TP53 by competing with the RBP HuR for interaction with TP53 mRNA (24). LncRNAs can also form partial hybrids with mRNAs and thereby control mRNA turnover and translation. Together, these findings indicate that lncRNAs regulate gene expression by different mechanisms and are thus capable of regulating cellular processes like differentiation, proliferation, stress response, and senescence (25-27). Accordingly, they also influence pathologies including cardiovascular disease, cancer, diabetes, AIDS, and neurodegeneration (28).
There is growing interest in the function of lncRNAs in cellular senescence. An earlier survey of senescence-associated lncRNAs (SAL-RNAs) was conducted in replicatively senescent WI-38 fibroblasts. This study identified senescence-regulatory lncRNAs; for example, reduction of SAL-RNA1 enhanced the appearance of senescence traits (29). Other SAL-RNAs, including TERC, HOTAIR, MALAT1, PINT, MEG3, ANRIL, Gadd7, 7SL, UCA1 and PANDA, have been functionally linked to senescence (24,26,30). However, little is known about the lncRNAs that might be implicated in regulating OIS. Using RNA-seq analysis, Li et al., have recently reported altered expression of lncRNAs upon HRASG12V induced-senescence in BJ fibroblasts. Among them, lncRNA-OIS1 is upregulated during OIS and its silencing using shRNA selectively enhanced cell proliferation and reduced SA-β-gal activity (31).
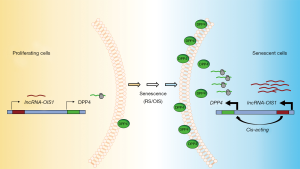
To understand the mechanism through which lncRNA-OIS1 regulates OIS, the authors analyzed gene expression profiles and found that cell cycle-related genes were highly enriched in lncRNA-OIS1-depleted cells. In situ hybridization (ISH) analysis indicated that lncRNA-OIS1 is localized both in the nucleus and the cytosol, indicating that it may regulate gene expression in both compartments. As highlighted above, lncRNAs can regulate transcription in cis by influencing transcription in the vicinity of the locus from which they are transcribed or in trans by influencing transcription at a distant locus. Global run-on sequencing (GRO-seq) analysis revealed enhanced transcription of lncRNA-OIS1 and the nearby gene DPP4 (dipeptidyl peptidase 4, also known as CD26) upon OIS, while silencing lncRNA-OIS1 lowered DPP4 mRNA production. DPP4 is an integral transmembrane glycoprotein that is widely expressed in several tissues (32). DPP4 is linked to cardiovascular diseases, metabolic diseases and cancer (33-35). It is involved in Type II diabetes mellitus (T2D), as it functions in the degradation of incretins such as glucagon-like peptide-1 (GLP-1); accordingly, a DPP4 inhibitor was developed to treat T2D and maintain insulin by preventing the degradation of incretins (36). While the specific mechanisms whereby DPP4 influences senescence are unknown, DPP4 was found to be highly abundant on the cell surface of senescent WI-38 cells and was used to target senescent cells using the antibody-dependent cell-mediated cytotoxicity (ADCC) methodology (37). Despite the robust increase of DPP4 mRNA levels in senescent cells (33), the molecular regulators of this rise were unknown until lncRNA-OIS1 was reported by Li et al., (31). The notion that DPP4 was a key effector of the lncRNA-OIS1-elicited senescence was supported by the fact that silencing DPP4 restored senescence even if lncRNA-OIS1 was silenced (31). These findings indicate that lncRNA-OIS1 rises during OIS and transcriptionally induces DPP4, which becomes a major effector of the ensuing senescent program.
While the full set of DPP4 transcriptional regulators is unknown, the identification of lncRNA-OIS1 is a major step forward, paving the way for the discovery of associated transcription factors. Since DPP4 expression was found elevated in other senescent models, including replicative and premature senescence, we hypothesize that lncRNA-OIS1 may drive DPP4 induction under different senescent triggers (Figure 1). It will be interesting to investigate if lncRNA-OIS1 also regulates the transcription of other RNAs upregulated in senescence, both coding and noncoding. The function of lncRNA-OIS1 in senescence in vivo also warrants analysis. With rising interest in devising approaches to eliminate senescent cells due to their harmful impact in older age, targeting lncRNA-OIS1 could have therapeutic benefits, possibly reducing damaging senescence-associated processes like inflammation. Finally, since DPP4 is further linked to diseases like T2D, there could be additional therapeutic value in targeting lncRNA-OIS1 in diabetes.
Acknowledgments
Funding: This work was funded in its entirety by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Jin Li (Cardiac Regeneration and Ageing Lab, School of Life Sciences, Shanghai University, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.01.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- He SH, Sharpless NE. Senescence in Health and Disease. Cell 2017;169:1000-11. [Crossref] [PubMed]
- Lopez-Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell 2013;153:1194-217. [Crossref] [PubMed]
- Herbig U, Ferreira M, Condel L, et al. Cellular senescence in aging primates. Science 2006;311:1257. [Crossref] [PubMed]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol 2013;75:685-705. [Crossref] [PubMed]
- Childs BG, Durik M, Baker DJ, et al. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med 2015;21:1424-35. [Crossref] [PubMed]
- Abdelmohsen K, Gorospe M. Noncoding RNA control of cellular senescence. Wiley Interdiscip Rev RNA 2015;6:615-29. [Crossref] [PubMed]
- Munk R, Panda AC, Grammatikakis I, et al. Senescence-Associated MicroRNAs. Int Rev Cell Mol Biol 2017;334:177-205. [Crossref] [PubMed]
- Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593-602. [Crossref] [PubMed]
- Zhu J, Woods D, McMahon M, et al. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 1998;12:2997-3007. [Crossref] [PubMed]
- Dimri GP, Itahana K, Acosta M, et al. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 2000;20:273-85. [Crossref] [PubMed]
- Lin AW, Barradas M, Stone JC, et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 1998;12:3008-19. [Crossref] [PubMed]
- Kuilman T, Michaloglou C, Mooi WJ, et al. The essence of senescence. Genes & Development 2010;24:2463-79. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 2013;9:e1003569 [Crossref] [PubMed]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science 2012;338:1435-9. [Crossref] [PubMed]
- Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014;26:10-8. [Crossref] [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010;329:689-93. [Crossref] [PubMed]
- Luo S, Lu JY, Liu L, et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016;18:637-52. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA (vol 147, pg 358, 2011). Cell 2011;147:358-69. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Seminars in Cell & Developmental Biology 2014;34:9-14. [Crossref] [PubMed]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Kang MJ, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res 2014;42:10099-111. [Crossref] [PubMed]
- Li J, Tian H, Yang J, et al. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biol 2016;35:459-70. [Crossref] [PubMed]
- Grammatikakis I, Panda AC, Abdelmohsen K, et al. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY) 2014;6:992-1009. [Crossref] [PubMed]
- Audas TE, Lee S. Stressing out over long noncoding RNA. Biochim Biophys Acta 2016;1859:184-91. [Crossref] [PubMed]
- Chen X, Yan CC, Zhang X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Briefings in Bioinformatics 2017;18:558-76. [PubMed]
- Abdelmohsen K, Panda A, Kang MJ, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell 2013;12:890-900. [Crossref] [PubMed]
- Ozes AR, Miller DF, Ozes ON, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016;35:5350-61. [Crossref] [PubMed]
- Li L, van Breugel PC, Loayza-Puch F, et al. LncRNA-OIS1 regulates DPP4 activation to modulate senescence induced by RAS. Nucleic Acids Res 2018;46:4213-27. [Crossref] [PubMed]
- Morimoto C, Schlossman SF. The structure and function of CD26 in the T-cell immune response. Immunol Rev 1998;161:55-70. [Crossref] [PubMed]
- Zhong J, Rao X, Rajagopalan S. An emerging role of dipeptidyl peptidase 4 (DPP4) beyond glucose control: potential implications in cardiovascular disease. Atherosclerosis 2013;226:305-14. [Crossref] [PubMed]
- Nargis T, Chakrabarti P. Significance of circulatory DPP4 activity in metabolic diseases. IUBMB Life 2018;70:112-9. [Crossref] [PubMed]
- Beckenkamp A, Davies S, Willig JB, et al. DPPIV/CD26: a tumor suppressor or a marker of malignancy? Tumour Biol 2016;37:7059-73. [Crossref] [PubMed]
- Duez H, Cariou B, Staels B. DPP-4 inhibitors in the treatment of type 2 diabetes. Biochem Pharmacol 2012;83:823-32. [Crossref] [PubMed]
- Kim KM, Noh JH, Bodogai M, et al. Identification of senescent cell surface targetable protein DPP4. Genes Dev 2017;31:1529-34. [Crossref] [PubMed]
Cite this article as: Munk R, Kim KM, Gorospe M, Abdelmohsen K. Senescence lncRNAs govern cell surface components: lncRNA-OIS1 transcriptionally elevates DPP4. Non-coding RNA Investig 2019;3:6.


