
Killing miR-softly: new clues to miRNA degradation by RNA targets
When talking about regulatory RNAs, and microRNAs in particular, attention should be paid to what types of molecules are present in the cell and in what numbers, rather than to their sizes. The distinction between small and long RNAs is based on RNA isolation protocols and is not a reflection of specific biogenesis pathways, regulatory mechanisms or biological roles. Conversely, sequence and quantity of expressed miRNAs are of paramount importance. As many other regulatory non-coding RNAs, miRNAs operate by interacting with other RNA molecules, typically mRNAs, at the level of the RNA Induced Silencing Complex (RISC), whose Argonaute proteins (Ago) act as essential catalytic subunits that mediate miRNA:target pairing. The miRNA:target interaction is based on a limited base-pair complementarity, which involves some nucleotides located at the 5′ end of the miRNA (the ‘seed’ region) as well as a complementary region usually located within the 3′ untranslated region of the target RNA (the miRNA responsive element, MRE). As it results into mRNA destabilization and protein synthesis inhibition, this type of miRNA:target pairing has become known as ‘miRNA directed target repression’ (1) (Figure 1A).
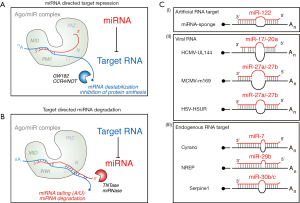
For a long time, this mechanism was believed to act unidirectionally, with most, if not all, cellular mRNAs being vulnerable to degradation by miRNAs. Once loaded into Ago, miRNAs are sheltered from the action of intra- and extracellular RNAses, thus usually behaving as extremely stable molecules with long half-lives (>24 h). When a series of separate reports showed that in specific cases, such as growth factor stimulation of fibroblasts (11-13) and exposure of neurons to light (14), some miRNAs are quickly down-regulated even in the absence of cell division, an active mechanism of degradation was suggested to exist. Indeed, it has now become common knowledge that a mechanism acting in the opposite direction to “miRNA directed target repression” exists, with some RNA targets evading silencing and inducing, instead, degradation of their cognate miRNAs. This mechanism is known as ‘target-directed miRNA degradation’ or TDMD (15) and relies on a specific miRNA:target architecture able to dislodge the miRNA tail (3′ end) out of the hydrophobic patch of Ago (PAZ domain) and expose it to a hydrophilic environment where it can be accessed by terminal nucleotidyl transferases (TNTases) or exonucleases (called ‘miRNases’) (Figure 1B). A typical hallmark of miRNA degradation is, indeed, the accumulation of certain miRNA isoforms (isomiRs) characterized by 3′ end modifications, such as the addition of non-templated nucleotides (tailing, usually A or U) or the shortening of the 3′ end (trimming) (11,15).
At first, target-directed miRNA degradation was observed in vitro (16) or induced in cells using the same synthetic targets commonly used to inhibit miRNA functions (‘miRNA sponges’) (Figure 1C) (2-4,15). Further clues came from studies on viruses able to repress host miRNAs for their own survival and propagation. In particular, Joan Steiz’s lab showed that Herpesvirus saimiri (HSV) HSUR RNA induces degradation of miR-27a/b in a sequence-specific and binding-dependent manner (5). Other cases of viral RNAs able to induce degradation of host miRNAs were documented (Figure 1C) (6,7) and additional studies on miRNA degradation highlighted an intriguing parallelism between miRNA decay dynamics and the expression of targets with extended complementarity (11). Finally, confirmation of the physiological relevance of TDMD came last year with a series of independent studies on endogenous RNAs able to trigger degradation of their cognate miRNAs in mammalian cells (Figure 1C): (I) Serpine1, shown to trigger miR-30b/c degradation in mouse (10); (II) libra/Nrep, able to degrade miR-29b in zebrafish and mouse, respectively (9); and (III) Cyrano, capable of inducing miR-7 decay in mouse (8). In each one of these studies, new and important aspects of the TDMD mechanism emerged. Nrep and Cyrano are almost exclusively expressed in the brain (where TDMD is very effective), thus conferring a spatial restriction to miRNA expression. Serpine1 is transcribed in fibroblasts, where it induces rapid miRNA degradation in quiescent cells stimulated by serum (temporal restriction). Therefore, TDMD seems to potentially function in every tissue although in neuronal cells it might work with higher processivity than in non-neuronal cells. Both Nrep and Serpine1 target specific members that belong to large miRNA families (the miR-27 family has 3 members and the miR-30 family 5) and multi-gene transcriptional clusters (miR-27b is transcribed with -23b and -24, miR-30b with -30d, and miR-30c with -30a, -30d and -30e). Hence, TDMD exerts a spatiotemporal control over miRNA levels and increases the flexibility of miRNA expression by uncoupling co-transcriptional regulation. Importantly, TDMD appears to be unrelated to the coding function of cellular RNAs. Removal of the MRE from both Nrep and Serpine1 abolished miRNA degradation by TDMD, but had no major effects on protein or mRNA levels. Indeed, both libra (in zebrafish) and Cyrano (in mouse) are actually lncRNAs with no reported coding function. Therefore, TDMD might be considered a non-coding mechanism shared by both coding and non-coding transcripts. Endogenous targets involved in TDMD are able to influence gene expression in trans by interfering with miRNA-mediated repression. Disruption of Cyrano induced an increase in miR-7 target repression in the brain and in the skeletal muscle. Loss of the Serpine1:miR-30b/c interaction (MRE knock-out) increased repression of miR-30 targets, deeply changed the expression of genes upon serum stimulation and induced cellular phenotypes related to cell cycle and apoptosis. Loss of the Nrep:miR-29b interaction generated an in vivo phenotype characterized by impaired motor functions in fish and mice, similar to the one resulting from the loss of the entire genetic locus (Nrep knock-out), thus suggesting a relevant role for TDMD in animal behaviour.
How many endogenous TDMD targets are there? How can they be identified? How exactly does TDMD work? These are the unsolved questions and future challenges we are faced with. Understanding the mechanistic details of miRNA degradation and TDMD will not only allow us to make new discoveries in the field of fundamental research, but it will also open new opportunities for the application of miRNA-based therapeutics to human disease.
Acknowledgments
Funding: The author is grateful for funding from Associazione Italiana per la Ricerca sul Cancro (AIRC, IG14085 and IG18774) and Cariplo (2015-0590) and thanks Claudia Crovace for manuscript editing.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Jing Shi (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.01.03). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Baccarini A, Chauhan H, Gardner TJ, et al. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol 2011;21:369-76. [Crossref] [PubMed]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007;4:721-6. [Crossref] [PubMed]
- Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 2009;6:63-6. [Crossref] [PubMed]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010;328:1563-6. [Crossref] [PubMed]
- Lee S, Song J, Kim S, et al. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 2013;13:678-90. [Crossref] [PubMed]
- Marcinowski L, Tanguy M, Krmpotic A, et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog 2012;8:e1002510 [Crossref] [PubMed]
- Kleaveland B, Shi CY, Stefano J, et al. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018;174:350-62.e17. [Crossref] [PubMed]
- Bitetti A, Mallory AC, Golini E, et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 2018;25:244-51. [Crossref] [PubMed]
- Ghini F, Rubolino C, Climent M, et al. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat Commun 2018;9:3119. [Crossref] [PubMed]
- Marzi MJ, Ghini F, Cerruti B, et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Marzi MJ, Puggioni EM, Dall'Olio V, et al. Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. The Journal of cell biology 2012;199:77-95. [Crossref] [PubMed]
- Rissland OS, Hong S-J, Bartel DP. MicroRNA Destabilization Enables Dynamic Regulation of the miR-16 Family in Response to Cell-Cycle Changes. Molecular Cell 2011;43:993-1004. [Crossref] [PubMed]
- Krol J, Busskamp V, Markiewicz I, et al. Characterizing Light-Regulated Retinal MicroRNAs Reveals Rapid Turnover as a Common Property of Neuronal MicroRNAs. Cell 2010;141:618-31. [Crossref] [PubMed]
- de la Mata M, Gaidatzis D, Vitanescu M, et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep 2015;16:500-11. [Crossref] [PubMed]
- Ameres SL, Horwich MD, Hung JH, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010;328:1534-9. [Crossref] [PubMed]
Cite this article as: Nicassio F. Killing miR-softly: new clues to miRNA degradation by RNA targets. Non-coding RNA Investig 2019;3:5.


