Emerging role of long noncoding RNAs and circular RNAs in pancreatic β cells
Introduction
Diabetes mellitus affecting more than 400 million people across the globe is one of the major health challenges today (1). Diabetes is a metabolic disorder characterized by dysfunctional production or sensing of insulin, leading to high glucose levels in the blood. Diabetes mellitus is categorized into Type 1 diabetes mellitus (T1DM) and Type 2 diabetes mellitus (T2DM), defined by the deficiency of insulin production by pancreatic β cells and by the loss of insulin receptor sensitivity on the target cells respectively (2,3). T1DM is an autoimmune disease leads to the destruction of pancreatic β cells accounts for less than 10% of the diabetic population while the rest 90% is T2DM. Glucose homeostasis in the body is regulated by the hormone insulin, which is secreted by pancreatic β cells (4-6). Increased extracellular glucose triggers a rapid upregulation of insulin secretion from the insulin storage granules and the secreted insulin is replenished by increased insulin biosynthesis through the activation of diverse gene regulatory mechanisms (5,7,8). In early stages of the T2DM, β cells increase their mass and insulin production to maintain glucose levels and overcome insulin resistance (9,10). However, long-term hyperglycemia leads to β cell failure by various mechanisms, including endoplasmic reticulum stress, mitochondrial dysfunction, and glucolipotoxicity (10). Most of the studies focused on elucidating the role of key protein-coding genes that are involved in insulin expression, secretion, and β cell apoptosis. However, the protein-coding sequences account for only 2% of the human genome (11,12). Interestingly, more than 85% of the human genome is likely to be transcribed and the majority of the transcripts are noncoding (nc) RNAs in nature (11-13). The ncRNAs include ribosomal (r)RNAs, transfer (t)RNAs, micro(mi)RNAs, piwi-interacting (pi)RNAs, small nuclear (sn)RNAs, long noncoding (lnc)RNAs and circular (circ)RNAs (14-17). In fact, we now know that these ncRNAs play a key role in regulating various steps of gene expression. While classical ncRNAs like rRNAs, tRNAs, snRNAs, and miRNAs are well characterized for their function in gene regulation, lncRNAs and circRNAs are poorly characterized (14-16,18-22). In this review, we will discuss the function of lncRNAs and circRNAs in insulin production.
LncRNAs are a heterogeneous class of >200 nucleotides linear transcripts that lack the protein-coding ability (11,23). The Encyclopedia of DNA Elements (ENCODE) Project Consortium revealed that more than 28,000 distinct lncRNA are transcribed from the human genome (11). LncRNAs are known to regulate every step of gene expression, including transcription, pre-mRNA splicing, mRNA stability, and translation (19,24-26). LncRNAs may act as a decoy for transcription factors, as sponges for miRNAs and RNA-binding Proteins (RBPs), as host genes for miRNAs, as scaffolds for protein complexes, and as stabilizer/destabilizer of mRNAs to regulate gene expression (25). A few lncRNAs are reported to associate with polyribosomes and translate into peptides (27). Various key cellular events like cell proliferation, growth, senescence, differentiation, and secretion are known to be regulated by lncRNAs. LncRNAs have been functionally associated with several diseases including cancer, nervous disorder, cardiovascular disease, muscular dystrophy, and diabetes (28-31).
Another class of novel ncRNA called circRNA was initially discovered in 1980s in plant viroid (32). Later few circRNAs were found to be expressed in eukaryotic cells and thought to be generated from erroneous splicing (33-36). Recently, high-throughput RNA-sequencing and unbiased analysis of mammalian transcriptome identified thousands of endogenous covalently closed loop circRNAs (37-39). CircRNAs are abundant, ubiquitously expressed, conserved, and show tissue-specific expression pattern (37,39-41). CircRNAs are shown to be originated from the pre-mRNAs by a process called backsplicing where an upstream 5’ splice site is ligated to a downstream 3’ splice site by head-to-tail nonlinear splicing (37,42,43). Although little is known about its function, increasing pieces of evidence established that they may act as miRNA sponges, act as a decoy for RBPs, compete with linear splicing, and translated into proteins (21,44). Although a numerous number of circRNAs have been identified to date, few circRNAs were reported to be involved in various diseases including cancer, muscular dystrophy, and diabetes (45). The role of lncRNAs and circRNAs in diabetes is not yet well understood but there is increasing evidence about their involvement in the development of diabetes.
Characteristics and function of lncRNAs
The protein-coding mRNAs represent less than 5% of the whole transcriptome while the rest are ncRNAs (16). The ncRNAs are broadly divided into short ncRNAs which are less than 200 nucleotides (nt) and long ncRNA with a length of more than 200 nt. Development in the next-generation sequencing (NGS) approaches revealed the expression of thousands of lncRNAs with no apparent protein-coding role. Recent ENCODE project revealed that the number of lncRNAs expressed from the human genome is more than the number of protein-coding genes (11). LncRNAs are mainly transcribed by RNA Polymerase (Pol) II or III in humans (46). Epigenetic modifications like H3K4me3 and H3K36me3 are associated with lincRNA transcription by RNA Pol II while H3K4me1/2/3 or H3K4ac are associated with Pol III transcription (46-48). Although it’s difficult to classify the lncRNAs, they are broadly classified into sense overlap, antisense overlap, bidirectional, intronic lncRNAs, and intergenic lncRNAs depending on their location relative to the nearest protein-coding gene (49). Many lncRNAs are low abundant and not well conserved during evolution, posing a challenge for their functional characterization (50). However, some of the lncRNAs are ultra-conserved in various organisms ranging from Xenopus and chicken to human (11,51). Although lncRNA sequences are not well conserved, the lncRNA promoters show higher conservation similar to the mRNA promoters suggesting their regulated expression (11). LncRNAs are differentially localized into distinct subcellular compartments like nucleus, cytoplasm, and mitochondria to execute different activities. The nuclear lncRNAs can regulate transcription and splicing while the cytoplasmic lncRNAs can regulate posttranscriptional events such as mRNA stability, translation, protein localization, and protein modification (25).
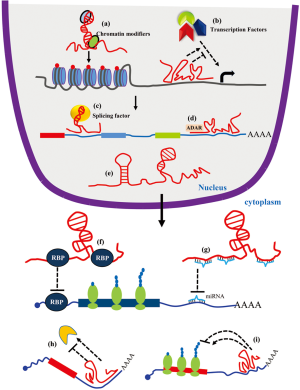
For a long time, lncRNAs were thought to be non-functional transcripts generated by transcriptional errors and termed as junk RNA. Recent development in transcriptomic moved the lncRNAs to the forefront of RNA research. Although the function of most of the identified lncRNAs is unknown, some of the lncRNAs are very well characterized. LncRNAs can regulate gene expression by interacting with diverse molecules and forming macromolecular complexes. As shown in Figure 1, lncRNAs can regulate transcription by recruiting chromatin modifiers to specific genes or by interacting with various transcription factors (24,52). Some lncRNAs can directly interact with the pre-mRNAs or splicing factors to regulate pre-mRNA splicing (53). Moreover, lncRNAs also recruit RNA modifiers like ADAR and regulate RNA modifications (54). LncRNAs can act as a decoy for proteins and inhibit the protein from interacting with its target mRNA or modify catalytic activity of the protein (19,25). Additionally, the presence of miRNA target sites on lncRNAs makes them a competitive endogenous (ce)RNA for the miRNAs and inhibit miRNA activity by its sponging ability (19,25). Interestingly, direct interaction of lncRNAs with translating mRNAs can alter the stability and/or translation of the target mRNA (19).
LncRNAs regulate β cell function
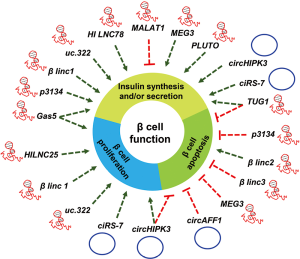
A major portion of the genome is transcribed as ncRNAs and many of them are reported to regulate gene expression. LncRNAs are shown to regulate gene expression at the transcriptional and posttranscriptional level. Indeed, lncRNAs are reported to play critical role in various biological processes including cell proliferation, differentiation, senescence and aging (28,29,55,56). Although many studies suggest the important role of lncRNA in development and disease, few studies investigated the functional significance of lncRNAs in the context of pancreatic β cell function and diabetes. Many lncRNAs are reported to be expressed in cell-type specific manner in islet β-cells and are termed as islet lncRNAs. In this section, we describe and discuss the emerging role of lncRNAs in pancreatic β cells (Table 1).
Table 1
| LncRNA/circRNA | Aliases | Overlapping/closest protein-coding gene | Target | Function | Reference | ||
|---|---|---|---|---|---|---|---|
| β cell proliferation | β cell apoptosis | Insulin biosynthesis and/or secretion | |||||
| MALAT1 | HCN, LINC00047 | – | miR-17, TXNIP | – | – | ↓ | (57) |
| GAS5 | NCRNA0003, SNHG2 | – | PDX1, MAFA | ↑ | – | ↑ | (58) |
| HI-LNC25 | LINC01370, HILNC25 | – | GLIS3 | ↑ | – | (59) | |
| HI-LNC78 | Tunar | – | – | – | – | ↑ | (60) |
| PLUTO | HI-LNC71, PDX1AS1, PLUT | PDX1 | PDX1 | – | – | ↑ | (60) |
| βLINC1 | HI-LNC15 | – | Nkx2.2 | ↑ | – | ↑ | (61) |
| βlinc2 | – | – | ___ | – | ↑ | – | (62) |
| βlinc3 | – | – | – | – | ↓ | – | (62) |
| TUG1 | ENSG00000253352 | – | caspase-3/9, PDX1, MAFA | ↓ | ↑ | (63) | |
| MEG3 | Gtl2, FP504, LINC00023, ENSG00000214548, PRO0518, PRO2160 | – | PDX1, MAFA | ↓ | ↑ | (64) | |
| LncRNA-p3134 | ENST00000545923 | – | PDX1, MAFA, GLUT2 | ↓ | ↑ | (65) | |
| LncRNA uc.322 | – | SOX6 | PDX1, FOXO1, SOX6 | ↑ | – | ↑ | (66) |
| ciRS-7/CDR1as | hsa_circ_0001946 | CDR1 | miR-7, Myrip, |
↑ | – | ↑ | (67,68) |
| circHIPK3 | hsa_circ_0021592 | HIPK3 | miR-124, miR-338 | ↑ | ↓ | ↑ | (68) |
| circAFF1 | – | AFF1 | – | – | ↓ | – | (68) |
MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the best-characterized lncRNAs for its molecular function and diseases association. Expression levels of thioredoxin‐interacting protein (TXNIP) and MALAT1 are upregulated while mafA and miR‐17 are downregulated in MIN6 cells with cigarette smoke extract (CSE) (57). TXNIP controls the redox state of the cell by inhibiting thioredoxin and reported to inhibit insulin production in β cells (69). Interestingly, silencing of MALAT1 upregulates the expression of miR-17 and miR-17 is known to promote insulin production. MALAT1 silencing in β cells increases miR‐17 expression leading to suppression of TXNIP and increased insulin production (57). Intriguingly, diabetic patients who smoked shows a high level of MALAT1 and low level of miR‐17 in the serum compared to non-smokers. In sum, CSE induces MALAT1 expression that reduces miR-17 levels leading to upregulation of TXNIP which in turn inhibits insulin production in β‐cells of smoking individuals.
GAS5
Growth arrest specific transcript 5 (GAS5) is a well-characterized lncRNA, known to generate many small nucleolar (sno)RNAs (70). As the name indicates GAS5 is known to regulate cell proliferation and growth. The serum level of GAS5 in diabetic patients is significantly lower compared to non-diabetic one (71). Gas5 is abundantly expressed in normal pancreas while in db/db mice Gas5 is significantly downregulated (58). In Min6 cells, silencing of Gas5 promotes cell cycle arrest at G1 and decreases insulin biosynthesis and secretion. Additionally, Gas5 knockdown in islets reduces the expression of insulin as well as transcription factors like Pdx1 and MafA underscore its role in insulin production and β cell function (58).
HI-LNC25
HI-LNC25 is a multiexonic lncRNA transcribed from the islet-specific active chromatin domain. It is specifically expressed in pancreatic β cells while the MAFB, the closest protein-coding gene localized in the same genomic loci is expressed in several tissues along with its abundant expression in β cells (59). Overexpression of HI-LNC25 by lentivirus in human β cells promotes the expression of GLIS3 that encodes an islet-specific transcription factor. Although silencing of HI-LNC25 does not affect glucose-induced insulin secretion, it downregulates GLIS3 mRNA levels (59). Interestingly, GLIS3 is involved in β cell growth and proliferation in response to insulin resistance. Thus, HI-LNC25 is a regulator of GLIS3 which regulates the development of type 2 diabetes.
HI-LNC78
HI-LNC78 is another islet lincRNA orthologous to mouse lncRNA Tunar is regulated by glucose (60). Silencing of HI-LNC78 reduces insulin content and show decreased glucose-stimulated insulin secretion in human EndoC-bH3 cells. Silencing of HI-LNC78 regulates the expression of various genes in β cells which is highly correlated with changes in gene expression by islet-specific transcription factors like HNF1A and MAFB suggests its role in β cell function (60).
PLUTO
PLUTO (PDX1 locus upstream transcript), also known as HI-LNC71 is an abundant antisense transcript lncRNA located 3 kb upstream of PDX1 gene. PDX1 is a well-known transcription factor involved in pancreas development and β cell differentiation. In T2DM patients or patients with glucose intolerance, PLUTO and PDX1 are significantly downregulated (60). Interestingly, the decrease in PLUTO expression alters chromatin architecture reducing the association of PDX1 promoter with its enhancer resulting in reduced expression of PDX1 (60). These data highlight the role of lncRNAs in the regulation of β cell development and function.
βLINC1
βLINC1 (β-cell long intergenic noncoding RNA 1), a 6.8 kb long lncRNA located nearly 20kb upstream of the transcription factor called NKX2.2 (61). In mammals, it is highly conserved and show islet specific expression pattern similar to other islet lncRNAs. The orthologous mouse βlinc1 has no protein coding ability. In mouse H3K27ac & H3K4me1/3 modifications are enriched around βlinc1 genomic region and the binding of transcription factors like Foxa2, NeuroD1, and Pdx1 to the putative βlinc1 promoter region is evident (61). The expression of βlinc1 is restricted to trunks and islets of the developing pancreas and it is mostly localized in the nucleus suggesting its role in transcriptional regulation. Knockdown of βlinc1 reduces the expression of Nkx2.2 which is responsible for transcription of many genes involved in β cell maturation and function. Interestingly, βlinc1 knockout mice show abnormal insulin secretion in low glucose and displays elevated glucose intolerance (61). Silencing of βlinc1 in MIN6 cells upregulates the expression of somatostatin and suppresses the expression of Scg5 and Pax6 further indicates the involvement of βlinc1 in β cell function.
βlinc2 and βlinc3
Human lincRNA βLINC3 expression is downregulated in T2DM patients compared to normal donors and its expression is negatively correlated with BMI of the patients (62). Two mouse orthologue lncRNAs βlinc2 and βlinc3 are reported to be differentially expressed in the islets of diet-induced obese mice (62). βlinc3 is localized mostly in the nucleus while βlinc2 is present in both nucleus and cytoplasm. Tissue specific analysis of βlinc2 and βlinc3 found them to be expressed mostly in pancreatic islets compared to other tissues. The expression level of βlinc2 is positively correlated with body weight, insulinemia, and glycemia while the level of βlinc3 expression is negatively correlated. Interestingly, upregulation of βlinc2 or downregulation of βlinc3 promotes apoptosis in MIN6 cells without affecting the insulin secretion suggests their role in development of T2DM (62).
TUG1
Taurine upregulated gene 1 (TUG1) is a multiexonic, spliced, polyadenylated lncRNA, highly conserved in mammals but absent in other vertebrates. It is highly expressed in pancreas compared to other organs and its expression is downregulated with glucose treatment (63). Low level of TUG1 expression in NOD mice compared to BALB/c suggests its strong association with diabetes. TUG1 knockdown induces β cell apoptosis evidenced by increased expression of apoptosis-related proteins such as caspase-3, caspase-9, GLUT2, Pdx1, MafA and decreased expression of antiapoptotic protein bcl-2 (63). Furthermore, silencing of TUG1 reduces insulin biosynthesis and secretion suggesting a definitive role of TUG1 in β cell function.
MEG3
Maternally expressed gene 3 (MEG3) is an imprinted lncRNA known to be involved in neurogenesis and retinal development. Furthermore, the low level of Meg3 is associated with diseases like Huntington’s disease and cancer (72,73). Meg3 is also required for overall growth and survival of mice. Interestingly, Meg3 is very abundantly and specifically expressed in pancreatic β cells (64). Its expression in β cells is also regulated by the level of glucose. Silencing of Meg3 decreases mRNA and protein levels of Ins2, pdx1 and MafA underscore its role in insulin synthesis and secretion (64). Silencing of Meg3 promotes apoptosis by increasing Bax and caspase3 expression and reducing Bcl 2 levels supports its role in the development of β cells. Together, these findings indicate that Meg3 regulates insulin production from β cells.
LncRNA-p3134
LncRNA-p3134 (also known as ENST00000545923) is known to suppress the expression of HOMA-β, that is associated with beta cell function (65). LncRNA-p3134 expression is upregulated with glucose treatment (65). Interestingly, overexpression of lncRNA-p3134 by glucose in MIN6 cells induces the expression of transcription factors like Pdx-1 and MafA as well as upregulation of GLUT2 expression, a glucose transporter associated with glucose-stimulated insulin secretion. In db/db mice induction of lncRNA-p3134 results in upregulation of TCF7L2 expression (65). Furthermore, lncRNA-p3134 act as a protective lncRNA to prevent beta cell destruction and apoptosis.
LncRNA uc.322
LncRNA uc.322 is a 224 base pair lncRNA originates from exonic region of SOX6 (SRY related HMG box 6). LncRNA uc.322 is highly conserved in mammals and found to be abundantly expressed in pancreatic tissue compared to other tissues (66). Overexpression of lncRNA uc.322 upregulates the expression levels of PDX1 and FOXO1. Knockdown of lncRNA uc.322 in MIN6 cells reduces insulin secretion and ATP concentration while its overexpression has opposite effect. SOX family proteins especially SOX 4 is associated with beta cell growth and development whereas SOX 6 promotes secretion of insulin (66). Expression level of lncRNA uc.322 is positively correlated with SOX6 expression which suggest its role in beta cell development and insulin transcription.
Characteristics and functions of circRNAs
In the early 1980s, a novel class of covalently closed loop circRNAs was initially discovered in plant viroid (32). For a long time, this kind of molecules was considered as splicing artifacts. Their existence and functional significance were neglected until the recent development of NGS technologies (37,39). Unlike linear RNAs, circRNAs are identified by the presence of the backsplice sequence at the 5’ to 3’ ligation junction (74). Global analysis of circRNA expression relies on high-throughput RNA-sequencing (RNA-seq) followed by identification of non-linear backsplice reads using bioinformatics tools (75). CircRNA enrichment methods like depletion of linear RNA by RNase R and/or poly(A) RNA and rRNA depletion were developed for specific identification of circRNAs by RNA-seq, as non-linear reads may be generated by various mechanisms including backsplicing, template switching of RT or trans splicing (40,74-78). High throughput RNA-seq discovered more than hundred thousand circRNAs in humans (39,77-79). As many of the circRNAs are low in abundance, high sequencing depths of 50 million reads or more are required for their identification and quantification (75,78,80,81). Although RNA-seq is the most preferred method for circRNA identification, methods like microarrays, RT-PCR, and northern blot analysis are also used for checking their expression (39,74,82,83).
As the majority of the identified circRNAs are generated from the exons of mRNAs, the canonical splicing machinery is believed to be involved in the biogenesis of circRNAs (42,43,84). CircRNAs are generated from the pre-mRNAs by a process called backsplicing where a downstream 5’ splice donor reversely attacks an upstream 3’ splice acceptor site of pre-mRNA forming a covalently closed circRNA lacking the 5’ and 3’ ends (42,43). For multiexon circRNAs, backsplicing is often coupled with canonical splicing to remove the intervening introns generating the mature exonic (E)circRNAs (37-40). Although backsplicing is not that efficient compared to linear splicing, the canonical splicing signals and the speed of transcription by RNA pol II have been shown to regulate the biogenesis of circRNAs (43). Various RBPs and inverted repeat sequences in the flanking intron are also involved in the regulation of circRNA biogenesis by backsplicing (42,85,86). Very often, the formation of EcircRNA leads to skipping of those exons from the mature mRNAs leading to expression of splice variants. Some EcircRNAs retains the intervening intron and termed as Exon-Intron (EI)circRNAs (87). Many intronic lariats have been reported to be resistant to degradation by the debranching enzyme leading to the generation of stable circular intronic (ci)RNAs (88). A recent study identified thousands of circRNAs generated from the intronic sequences with unknown mechanism of biogenesis and termed as intronic (I)cicRNAs (78).
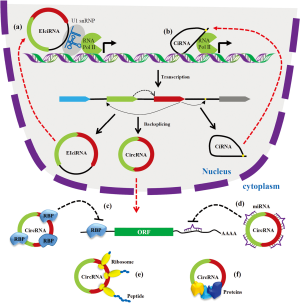
As shown in Figure 2, circRNAs generated from the pre-mRNA by backsplicing and regulate gene expression by various mechanisms. The EcircRNAs are localized in the cytoplasm and regulate posttranscriptional gene regulation (21,38). Many circRNAs harbor miRNA response elements (MRE) can act as miRNA sponges (89). CircRNAs with miRNA binding sites can act as an ceRNA and inhibit the miRNA from binding to its target mRNAs leading upregulation of target gene expression (21,89). CDR1as is one of the highly conserved and abundant circRNA discovered to contain more than 60 MREs for miR-7 (38,39). Accumulating studies have established that the circRNA-miRNA-mRNA network plays a critical role in gene expression (89,90). Besides miRNA sponging, circRNAs can act as a decoy for the RBPs and modulate the expression of the RBP target gene. For instance, circPABPN1 acts as a decoy for HUR and sequester HUR from binding to PABPN1 mRNA leading to decrease in PABPN1 translation (82). Another circRNA circMBL act as a sponge for the splicing factor MBL and regulate MBL expression (91). Various web tools have been developed to explore the interaction of circRNAs with miRNAs and RBPs (92-94). The ciRNAs and EIcircRNAs are localized in the nucleus and associate with RNA Pol II to regulate transcription of target genes (87). Although circRNAs are categorized as ncRNAs, a few circRNAs are reported to translate into peptides (95-97). Recent studies have shown that circRNAs can act biomarkers for human diseases (98). Although the function of some circRNAs have been characterized, the function of majority of circRNAs remains to be explored.
CircRNAs regulate β cell function
CircRNAs are emerging as novel regulators of gene expression and shown to be involved with disease pathogenesis. There is increasing evidence that circRNAs play critical roles in various pathophysiological conditions. Although some circRNAs are very well characterized and considered as important biological molecules for gene regulation, very little is known about their role in β cell function and diabetes. In this section, we review the functional significance of circRNAs in pancreatic β cells.
Cdr1as
Cdr1as (also termed as ciRS-7) is generated from antisense transcript of cerebellum degeneration-related antigen 1 (CDR1). Cdr1as was one of the first functional circRNAs identified to act as a sponge for miRNA. Interestingly, Cdr1as contains more than 60 binding sites for miR-7 leading to inhibition of miR-7 activity (39). Cdr1as is upregulated in mouse islets with forskolin and PMA treatment. As miR-7 is known to decrease insulin secretion, upregulation of Cdr1as promotes insulin secretion by inhibiting miR-7 activity (99). In fact, the myosin VIIA and Rab interacting protein (Myrip) and paired box 6 (Pax 6) are the direct targets of miR-7 (67). Myrip is known to be involve in secretory granules transportation and release, while Pax 6 is a transcription factor which binds to the promoter of ins1 and ins2 genes and leads to enhanced insulin biosynthesis and secretion. Expression of miR-7 inhibits Myrip and Pax 6 expression in mouse islets and MIN6 cells. Interestingly, overexpression of Cdr1as upregulates expression of Myrip and Pax 6 resulting in increased insulin transcription as well as increased insulin secretion in islets and MIN6 cells (67). Interestingly, Cdr1as expression is downregulated in the islets of db/db and ob/ob mice (68). Silencing of Cdr1as decreases the prolactin-stimulated proliferation of rat β cells and MIN6B1 cells. Together, Cdr1as regulate insulin biosynthesis and secretion by acting as a sponge for miR-7.
circHIPK3
CircHIPK3 is one of the most abundant circRNA expressed in a variety of cells including pancreatic β cells. CircHIPK3 is generated from the exon2 of HIPK3 mRNA by backsplicing. Reduction in circHIPK3 levels promotes apoptosis and reduces proliferation of β cells (68). In fact, prolactin fails to induce proliferation of circHIPK3 silenced MIN6B1 and β cells. Silencing of this circRNA reduces insulin mRNA levels as well as impairs glucose-stimulated insulin secretion. Modulation of circHIPK3 level impairs the function of the promoter region of insulin in MIN6B1 cells. Additionally, knockdown of circHIPK3 reduces insulin content in MIN6B1 cells suggesting that circHIPK3 plays a critical role at the level of translation or post-translation. The effect of circHIPK3 silencing on β cell function is also partly regulated through sponging miRNAs including miR-124-3p and miR-338-3p (68).
circAFF1
As the name indicates circAFF1 is produced from the AFF1 gene. CircAFF1 is one of the abundant circRNA expressed in pancreatic islets of human, mouse and rat as well as in insulin-secreting INS832/13 and MIN6B1 cell lines (68). CircAFF1 silencing induces apoptotic phenotype in MIN6B1 cells and primary rat β cells similar to apoptosis after treatment with proinflammatory cytokines. In contrast, silencing circAFF1 in MIN6B1 cells and of primary rat β-cells has no effect on cell proliferation and no effect on insulin biosynthesis or insulin secretion (68).
Concluding remarks and perspectives
In summary, with the development of various genomic technologies, a numerous number of lncRNAs and circRNAs are identified in various organisms. Many of the lncRNAs and circRNAs regulate gene expression by controlling gene transcription, pre-mRNA splicing, mRNA stability, and translation. Increasing pool of data indicates the crucial role of lncRNAs and circRNAs in development and diseases. Recent studies have focused on the use of lncRNAs and circRNAs as biomarkers for detection and prognosis of diseases. Increasing evidence showed that lncRNAs and circRNAs are involved in the pancreatic β cell physiology and development of diabetes. In this review, we discussed the emerging functional roles of lncRNAs and circRNAs associated with the development of diabetes focusing on pancreatic β cell function (Figure 3).
Without any doubt, lncRNAs and circRNAs are emerging as critical tools for diseases diagnosis and treatment. However, our current knowledge of lncRNAs and circRNAs in β cell function remains rather limited. Although functional role of various lncRNAs and circRNAs have been characterized, vast majority of lncRNAs and circRNAs remains to be explored in the context of diabetes. To gain insight into function of lncRNAs and circRNAs in development of diabetes, few major issues need to be addressed; (I) development of NGS with computational algorithms for identification and functional analysis is essential; (II) as lncRNAs and circRNAs regulate gene expression by interacting with DNA, RNA, and proteins, computational algorithms and integrated datasets needs to be developed to understand the complex crosstalk of regulatory molecules in gene regulation; (III) we need to develop suitable animal models to study the specific effect of lncRNA and circRNAs in diabetes. With these technological advancements, we expect to gain a deeper understanding of the role of lncRNAs and circRNAs in β cell function which may lead to development of effective therapies for diabetes.
Acknowledgments
AC Panda was supported by the Science & Engineering Research Board, a statutory body of the Department of Science &Technology (DST), Government of India. D Das, A Das, and AC Panda were supported by the Institute of Life Sciences Intramural Research Program, Department of Biotechnology, Government of India.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.11.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cho NH, Shaw JE, Karuranga S, et al. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract 2018;138:271-81. [Crossref] [PubMed]
- American Diabetes Associaiton. Classification and diagnosis of diabetes. Diabetes Care 2015;38:S8-16. [Crossref] [PubMed]
- Klöppel G, Lohr M, Habich K, et al. Islet pathology and the pathogenesis of type 1 and type 2 diabetes mellitus revisited. Surv Synth Pathol Res 1985;4:110-25. [PubMed]
- Banting FG, Best CH, Collip JB, et al. Pancreatic Extracts in the Treatment of Diabetes Mellitus. Can Med Assoc J 1922;12:141-6. [PubMed]
- Laville M, Nazare JA. Diabetes, insulin resistance and sugars. Obes Rev 2009;10:24-33. [Crossref] [PubMed]
- Rayburn WF. Diagnosis and classification of diabetes mellitus: highlights from the American Diabetes Association. J Reprod Med 1997;42:585-6. [PubMed]
- Docherty K, Clark AR. Nutrient regulation of insulin gene expression. FASEB J 1994;8:20-7. [Crossref] [PubMed]
- Plum L, Belgardt BF, Bruning JC. Central insulin action in energy and glucose homeostasis. J Clin Invest 2006;116:1761-6. [Crossref] [PubMed]
- Cnop M, Welsh N, Jonas JC, et al. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes 2005;54:S97-107. [Crossref] [PubMed]
- Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest 2006;116:1802-12. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760-74. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol 2014;49:16-32. [Crossref] [PubMed]
- Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016;937:3-17. [Crossref] [PubMed]
- Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet 2015;6:2. [Crossref] [PubMed]
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 2017;242:1136-41. [Crossref] [PubMed]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77-94. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013;425:3723-30. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Panda AC, Grammatikakis I, Munk R, et al. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA 2017;8. [PubMed]
- Panda AC, Sahu I, Kulkarni SD, et al. miR-196b-mediated translation regulation of mouse insulin2 via the 5'UTR. PLoS One 2014;9:e101084 [Crossref] [PubMed]
- Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3' UTRs. RNA 2012;18:825-43. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Bánfai B, Jia H, Khatun J, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 2012;22:1646-57. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Hagan M, Zhou M, Ashraf M, et al. Long noncoding RNAs and their roles in skeletal muscle fate determination. Noncoding RNA Investig 2017;1. [PubMed]
- Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737-50. [Crossref] [PubMed]
- Quan Z, Zheng D, Qing H. Regulatory Roles of Long Non-Coding RNAs in the Central Nervous System and Associated Neurodegenerative Diseases. Front Cell Neurosci 2017;11:175. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. [Crossref] [PubMed]
- Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986;323:558-60. [Crossref] [PubMed]
- Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993;73:1019-30. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Xia S, Feng J, Lei L, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 2017;18:984-92. [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [Crossref] [PubMed]
- Zhang Y, Xue W, Li X, et al. The Biogenesis of Nascent Circular RNAs. Cell Rep 2016;15:611-24. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Haque S, Harries LW. Circular RNAs (circRNAs) in Health and Disease. Genes (Basel) 2017;8. [PubMed]
- Barski A, Chepelev I, Liko D, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 2010;17:629-34. [Crossref] [PubMed]
- Sigova AA, Mullen AC, Molinie B, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A 2013;110:2876-81. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 2016;44:D203-8. [Crossref] [PubMed]
- Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635-40. [Crossref] [PubMed]
- Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008;454:126-30. [Crossref] [PubMed]
- Romero-Barrios N, Legascue MF, Benhamed M, et al. Splicing regulation by long noncoding RNAs. Nucleic Acids Res 2018;46:2169-84. [Crossref] [PubMed]
- Peters NT, Rohrbach JA, Zalewski BA, et al. RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts. RNA 2003;9:698-710. [Crossref] [PubMed]
- Grammatikakis I, Panda AC, Abdelmohsen K, et al. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY) 2014;6:992-1009. [Crossref] [PubMed]
- Abdelmohsen K, Panda A, Kang MJ, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell 2013;12:890-900. [Crossref] [PubMed]
- Sun Q, Xu H, Xue J, et al. MALAT1 via microRNA-17 regulation of insulin transcription is involved in the dysfunction of pancreatic beta-cells induced by cigarette smoke extract. J Cell Physiol 2018;233:8862-73. [Crossref] [PubMed]
- Jin F, Wang N, Zhu Y, et al. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic beta Cells. Cell Physiol Biochem 2017;43:2062-73. [Crossref] [PubMed]
- Morán I, Akerman I, van de Bunt M, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 2012;16:435-48. [Crossref] [PubMed]
- Akerman I, Tu Z, Beucher A, et al. Human Pancreatic beta Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab 2017;25:400-11. [Crossref] [PubMed]
- Arnes L, Akerman I, Balderes DA, et al. betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes Dev 2016;30:502-7. [Crossref] [PubMed]
- Motterle A, Gattesco S, Peyot ML, et al. Identification of islet-enriched long non-coding RNAs contributing to beta-cell failure in type 2 diabetes. Mol Metab 2017;6:1407-18. [Crossref] [PubMed]
- Yin DD, Zhang EB, You LH, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol Biochem 2015;35:1892-904. [Crossref] [PubMed]
- You L, Wang N, Yin D, et al. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J Cell Physiol 2016;231:852-62. [Crossref] [PubMed]
- Ruan Y, Lin N, Ma Q, et al. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet beta-Cell Function. Cell Physiol Biochem 2018;46:335-50. [Crossref] [PubMed]
- Zhao X, Rong C, Pan F, et al. Expression characteristics of long noncoding RNA uc.322 and its effects on pancreatic islet function. J Cell Biochem 2018;119:9239-48. [Crossref] [PubMed]
- Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. [Crossref] [PubMed]
- Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab 2018;9:69-83. [Crossref] [PubMed]
- Shalev A. Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Mol Endocrinol 2014;28:1211-20. [Crossref] [PubMed]
- Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 1998;18:6897-909. [Crossref] [PubMed]
- Carter G, Miladinovic B, Patel AA, et al. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin 2015;4:102-7. [Crossref] [PubMed]
- Sun L, Li Y, Yang B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53's transcriptional activity. Biochem Biophys Res Commun 2016;478:323-9. [Crossref] [PubMed]
- Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis 2012;46:245-54. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679-92. [Crossref] [PubMed]
- Suzuki H, Zuo Y, Wang J, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006;34:e63 [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Panda AC, De S, Grammatikakis I, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 2017;45:e116 [Crossref] [PubMed]
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666-70. [Crossref] [PubMed]
- Panda AC, Grammatikakis I, Kim KM, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res 2017;45:4021-35. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, De S, et al. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015;7:903-10. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361-9. [Crossref] [PubMed]
- Panda AC, Gorospe M. Detection and Analysis of Circular RNAs by RT-PCR. Bio Protoc 2018;8. [PubMed]
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103-11. [Crossref] [PubMed]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
- Li X, Liu CX, Xue W, et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell 2017;67:214-27.e7. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol 2018;1087:67-79. [Crossref] [PubMed]
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017;8:73271-81. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016;44:D209-15. [Crossref] [PubMed]
- Panda AC, Dudekula DB, Abdelmohsen K, et al. Analysis of Circular RNAs Using the Web Tool CircInteractome. Methods Mol Biol 2018;1724:43-56. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Das A, Gorospe M, Panda AC. The coding potential of circRNAs. Aging (Albany NY) 2018;10:2228-9. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267-74. [Crossref] [PubMed]
- Latreille M, Hausser J, Stutzer I, et al. MicroRNA-7a regulates pancreatic beta cell function. J Clin Invest 2014;124:2722-35. [Crossref] [PubMed]
Cite this article as: Das D, Das A, Panda AC. Emerging role of long noncoding RNAs and circular RNAs in pancreatic β cells. Non-coding RNA Investig 2018;2:69.