
Non-coding RNA in cardiovascular disease: a general overview on microRNAs, long non-coding RNAs and circular RNAs
Introduction
Cardiovascular diseases (CVDs) remain the main cause of morbidity and mortality in the Western world, and there is need for basic science research to provide insights into disease mechanisms. Indeed, obtaining a better understanding of the molecular and cellular mechanisms driving CVD development and progression is essential to identify new biomarkers and novel therapeutic targets in order to improve care and prevent the development of life-threatening complications.
Over the last decade, the advances in high-throughput sequencing technology have allowed the opportunity to expand our knowledge on the complexity of the human transcriptome, showing that the non-coding portion of the genome plays a more significant role in human biology than previously thought (1). Currently, we know that the most of the human genome is not translated into proteins, but transcribed into various classes of functional non-coding RNAs (ncRNAs) that are powerful regulators of a plethora of cellular and disease processes (2).
Based on their size, these molecules are classified into small ncRNAs (<200 nucleotides long), including microRNAs (miRNAs), and long ncRNAs (lncRNAs), exceeding a length >200 nucleotides. lncRNAs can also present circular form, called circular RNAs (circRNAs).
Recently, several review articles have been published discussing the involvement of three major types of ncRNAs (miRNAs, lncRNAs and circRNAs) in cardiovascular system, outlining their biogenesis, physiologic actions and pathogenic role (3-6). The present review discusses how ncRNAs (including miRNAs, lncRNAs and circRNAs) are involved in cardiovascular biology and diseases, highlighting their potential role as circulating diagnostic, and prognostic biomarkers and therapeutic targets. The review also addresses future directions in research, covering issues still unresolved and the relevant factors limiting their widespread use in the clinical practice.
miRNAs in the cardiovascular system
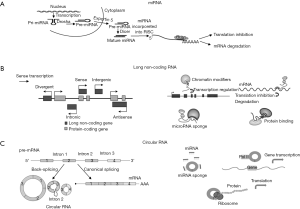
miRNAs are endogenous RNAs of ~22 nucleotides that negatively regulate expression of target genes by usually binding to the 3’ untranslated region (UTR) of mRNA and inhibiting their translation (7,8). They are synthesized as precursors in the nucleus (Figure 1), where they undergo maturation with several enzymatic reactions and are translocated to the cytoplasm where they exert their biological function recruiting specific silencing proteins that form the RNA induced silencing complex (RISC) (9,10). It has been predicted that, in humans, about 60% of mRNAs are targets for miRNAs and one miRNA may target more than 100 mRNAs (8).
Specific miRNAs are differently expressed in cardiac tissue and vascular cells, playing a crucial role as regulators of cardiovascular biological functions, including cardiovascular cell differentiation, growth, proliferation, apoptosis, angiogenesis and cell contractility (11).
Consequently, aberrant expression of miRNAs has been reported in heart suffering of several CVD, such as myocardial infarction (12) and end-stage cardiomyopathy (13).
Several miRNAs (miRNA-1, miRNA-133a, miRNA-208a/b, and miRNA-499) are believed to be cardiac-specific molecules and abundantly expressed in the myocardium (14).
In animal models, aberrant expression of cardiac-specific miRNAs has been associated with the onset and progression of cardiac conditions, such as arrhythmias, cardiac hypertrophy and fibrosis (15,16). The dysregulation of cardiac-specific miRNAs has been also reported in cardiac tissue of patients with heart failure and myocardial infarction (17,18).
However, other miRNAs (e.g., miRNA-21-5p, miRNA-126-3p) that are not cardiac-specific or muscle-enriched molecules are important players in several cardiovascular processes, contributing the onset and progression of CVDs (19).
Overall, expression profiling studies in experimental and human heart disease have shown that the expression of a large number of miRNAs is altered in several cardiovascular disorders (3-6), including myocardial infarction (miRNA-1, miRNA-20a, miRNA-21, miRNA-126, miRNA-155, miRNA-210, miRNA-214), cardiac arrhythmia (miRNA-1, miRNA-17-92, miRNA-106b-25, miRNA-133, miRNA-133a, miRNA-212), cardiac fibrosis (miRNA-21, miRNA-29, miRNA133), cardiac hypertrophy (miRNA-21, miRNA-23a, miRNA-24, miRNA-199, miRNA-208a) and heart failure (miRNA-1, miRNA-21, miRNA-29, miRNA-30, miRNA-195, miRNA-210, miRNA-499).
miRNAs are also critical in many key processes linked to vascular biology and atherosclerotic development, regulating endothelial dysfunction (miRNA-27b, miRNA-130a, miRNA-126, miRNA-221 and miRNA-222) and vascular smooth muscle cell proliferation and contractile function (miRNA-143 and miRNA-145) as well as inflammatory macrophage responses (miRNA-33, miRNA-155, miRNA-146a, miRNA-let7a, miRNA-21, miRNA-223 and miRNA-125a) (20-23).
Furthermore, a number of other specific miRNAs have been implicated in lipid metabolism and cholesterol homeostasis including miRNA-33 that is one of the most extensively studied miRNAs and it represses multiple genes involved in cellular cholesterol trafficking (24,25).
Recently, profiling analyses also reported differential expression of miRNAs (miRNA-21, miRNA-26a, miRNA-30b, miRNA-141, miRNA-125b, miRNA-148a, miRNA-204, miRNA-214) in cardiac valve disease, regulating key processes underlying disease progression, such as fibrosis, calcification, matrix degradation remodeling, and inflammation (26-28).
lncRNAs in cardiovascular physiopathology
lncRNAs are a heterogeneous group of RNA transcripts with lengths >200 nucleotides that can be classified as sense, antisense, intronic, intergenic and divergent lncRNAs according to their relative genome position (Figure 1). lncRNAs can be broadly classified into those that act in cis, influencing proximal events, and those that influence distal biological functions throughout the cell in trans (29,30).
Indeed, lncRNAs are involved in numerous and different biological events, such as chromatin structure changes, transcription and post-transcriptional processing, intracellular trafficking, and regulation of enzymatic activity (29,30). lncRNAs can also regulate the activity of other ncRNAs, specifically miRNAs, by acting as competing endogenous RNAs (31).
LncRNAs are less conserved than miRNAs, suggesting a species-specific role of these RNA molecules (32). Although the dysregulation of lncRNAs has been implicated in various human diseases (33), the functional roles and mechanisms of most lncRNAs remain, however, elusive (34).
LncRNAs have been reported to predominantly function as key regulators of cell fate commitments in embryonic and organism development (35). In 2013, a novel lncRNA, named Braveheart, was identified in mouse heart and demonstrated to be a key regulator of cardiovascular lineage and cardiac gene expression during heart development (36). Subsequently, an elegant study identified a human-specific lncRNA, named Heart Brake LncRNA 1 (HBL LncRNA 1), which negatively regulates human cardiomyocyte development from pluripotent stem cells (hiPSCs) by silencing miRNA-1 activity (37).
To date, the deregulation of lncRNAs has been reported in some cardiovascular conditions such as myocardium infarction, myocardial fibrosis, cardiac hypertrophy and heart failure (38-45).
For instance, lncRNA-Wisp2 super-enhancer-associated RNA (lncRNA-wisper) has been found to be a cardiac fibroblast-enriched lncRNA that regulates cardiac following myocardial infarction in a murine mode (46). Moreover, the expression of lncRNA-wisper was also correlated with the cardiac fibrosis in heart tissue from human patients suffering from aortic stenosis (46).
Myocardial infarction associated transcript (MIAT) was originally identified as a non-coding functional RNA able to confer risk of myocardial infarction (47). Subsequently, through a mouse model of myocardial infarction, it has been demonstrated the critical involvement in cardiac fibrosis and dysfunction (48), probably by sponging miRNA-150 (49) and miRNA-93 (50) expression in cardiomyocytes.
The lncRNAs cardiac hypertrophy-associated transcript (CHAST) is increased upon pressure overload-induced HF in mice and is preserved in humans. Interestingly, CHAST homolog in humans is significantly up-regulated in human embryonic stem cell-derived cardiomyocytes upon hypertrophic stimuli and in hypertrophic heart tissue from aortic stenosis patients (51). On the contrary, Myheart or Mhrt is a cardiac lncRNA, located in the locus of the cardiac-specific gene myosin heavy chain 7, which prevents cardiomyocyte hypertrophy by sequestering Brg1, a stress-activated, ATP-dependent chromatin-remodeling factor, and, therefore, avoiding the transcription of hypertrophy related genes that are induced during stress through a Brg1-mediated chromatin remodeling mechanism (52).
circRNAs in CVD
circRNAs are a peculiar group of lncRNAs, consisting of at least a few hundred nucleotides (53). As schematized in Figure 1, circRNAs are generated via back-splicing, a form of alternative splicing, and characterized by covalently closed loop structures through joining the 3’ and 5’ end together by exon or intron circularization (53,54). In the past, circRNAs were considered to have no biological function (55), but it has been recently demonstrated that they are abundant and preserved in mammalian cells and have biological functions by regulating gene expression at the transcriptional or post-transcriptional level (53,56,57).
circRNAs have the ability to bind to miRNAs and consequently regulate miRNA function, acting as sponge (58). Moreover, circRNAs have a relatively higher biological stability than linear RNA due to their circular structure that cannot be recognized or hydrolysed by RNA exonuclease (59).
Although several RNA-sequencing analyses have revealed that there is a high-abundance of specific cardiac-expressed circRNAs in human heart (60,61), much less is known about their role in diseased cardiac tissue. Recently, experimental studies have begun to delineate the role of circRNAs as crucial modulators of miRNA levels in cardiac conditions, such as myocardial infarction (62), cardiac fibrosis (63,64) and hypertrophy (65).
A circRNA profiling in left ventricle RNA samples with hypertrophic and dilated cardiomyopathy and unaffected heart tissues found 80 circRNAs expressed from the titin (TTN) gene (66). In particular, the authors showed that the RNA-binding motif protein 20 (RBM20), an important pathogenic gene of dilated cardiomyopathy, regulated the biosynthesis of circRNAs from the TTN gene (66).
Recently, a very interesting study identified an abundant expression of cardiac circRNAs (circSLC8A1, circCACNA1D, circSPHKAP and circALPK2) in heart tissues as well as in human induced pluripotent stem cells-derived cardiomyocytes, which might be used as biomarkers (67). Furthermore, the expression level of circSLC8A1 significantly increased in specimens from patients with dilated cardiomyopathy when compared to the healthy controls (67).
Altogether, these findings encourage future investigation to identify the differential expression circRNAs in different disease phenotypes in patients.
Circulating ncRNAs as biomarkers
In addition to the relevance of ncRNAs as regulators in the molecular mechanisms of disease, numerous evidence suggests their potential use as novel biomarkers for the diagnosis and clinical decision making (19,68,69). The best-studied circulating ncRNAs group is represented by circulating miRNAs, that are released into circulation usually packaged in different micro-particles (exosomes, micro-vesicles and apoptotic bodies) or associated with lipoprotein complexes or RNA-binding proteins, which provide stability and resistance to plasma RNase digestion and enable miRNA transfer from one cell to another (70).
Numerous studies have explored the potential of miRNAs as clinical biomarkers in CVD in the diagnosis and prognosis of CVDs (19,68-71).
For instance, a recurrent group of cardiomyocyte-enriched miRNAs (miRNA-1, miRNA-133, miRNA-208a/b and miRNA-499) and non-cardiac miRNAs (miRNA-21, miRNA-26a, miRNA-27a, miRNA-30c/d, miRNA-106a-5p, miRNA-122, miRNA-126, miRNA-134, miRNA-145, miRNA-146, miRNA-150, miRNA-197, miRNA-199, miRNA-223, miRNA-328, miRNA-423-5p, miRNA-486) in plasma or serum have been suggested as biomarkers of coronary artery disease and myocardial infarction as well as correlated with the diagnosis and the prognosis of heart failure (19,68-71).
Other circulating miRNAs (miRNA-1, miRNA-21, miRNA-133a/b, miRNA-146a, miRNA-150, miRNA-328) have been associated with cardiac arrhythmia and atrial fibrillation (19,69).
Other studies identified several circulating (miRNA-1, miRNA-21, miRNA-22, miRNA-133, miRNA-210, miRNA-382) as potential biomarkers for valvular heart disease, especially aortic stenosis, in combination with clinical and imaging parameters (26,72,73).
Despite these promising findings on circulating miRNAs as novel CVD biomarkers, great uncertainty remains on their diagnostic feasibility and clinical use due to inconsistent results among studies, attributable, at least in part; to a number of technical limitations for their measure in biological fluids (71).
In addition to miRNAs, lncRNAs can be released into the extracellular space and subsequently be detected in body fluids, such as serum and plasma (19,69).
Accordingly, some circulating lncRNAs were recently described as potential biomarkers for coronary artery disease/acute myocardial infarction (ZFAS1, UCA1, HOTAIR, LIPCAR, ANRIL, KCNQ1OT1, LncPPARδ*, CoroMarker) and heart failure (SENCR, NRON, LIPCAR, MHRT), encouraging future studies to determine the value of lncRNAs as novel cardiac biomarkers (38,69,74-79).
Moreover, circRNAs have a great potential as they are extraordinarily more stable in body fluids than other noncoding RNAs because their circularization protects them from endonuclease activities (80).
Several studies have also reported circRNAs as diagnostic and prognostic biomarkers of CVD (4,19). For instance, a clinical study identified a circRNA, MICRA, whose expression levels measured at reperfusion in peripheral blood samples of 642 patients with acute myocardial infarction from two independent cohorts, predicted left ventricular dysfunction after 3 to 4 months (81).
Furthermore, a circRNA, designated circRNA_081881, may be correlated with myocardial infarction since it was down‐regulated more than 10‐fold in blood samples of patients (82).
An increased risk of atherosclerosis has been associated with the circular isoform of ANRIL (cANRIL) that is associated with the INK4/ARF locus on human chromosome 9p21 (83).
A very recent study investigated the circRNA profile in the peripheral blood of patients with coronary artery disease, reporting a circRNA (hsa_circ_0124644) as a biomarker with a great diagnostic value for the disease (84).
The main circulating ncRNAs involved in the CVDs are summarized in Figure 2.

Concluding remarks and future perspectives
In conclusion, ncRNAs are ubiquitous RNA molecules that play a key role in modulating the molecular mechanisms underlying the pathogenesis of the CVD. Accordingly, there are attractive and promising applications of ncRNAs in the diagnosis and treatment of these diseases (3-6).
From a therapeutic perspective, the use of molecules to inhibit or overexpress small and long ncRNAs could be used as novel therapeutic strategy to combat various cardiac disorders.
For instance, pharmacological inhibition of miRNA-33a and miRNA-33b (miRNAs involved in the regulation of cholesterol transport) led to an increased levels of plasma HDL cholesterol and a coincident reduction of VLDL triglycerides without any adverse side effects in non-human primates, supporting the development of antagonists (also called antagomiRs or blockmiRs) of miRNA-33 as potential therapeutics for dyslipidemia and related atherosclerotic diseases (85).
Furthermore, inhibition of miRNA-29 in vivo abrogates aortic dilation in mice, suggesting that blocking miRNA-29 may represent a potential molecular target to treat aortic aneurysms (86).
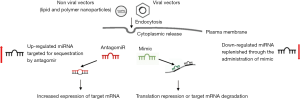
On the other hand, the administration of systemic miRNAs through miRNA mimics or introducing genes coding for miRNAs into viral constructs could be an attractive therapeutic approach for many diseases (87).
In the cardiovascular field, it has been recently shown that the overexpression with mimic (88) or adeno-associated virus-mediated cardiomyocyte-targeted expression of miR-378 (89) produced significant anti-apoptotic and anti-hypertrophic activities in cardiac cells, representing a potential treatment for ischemic heart disease. Additionally, a recent study demonstrated the feasibility of using viral-based delivery of DNA code for non-native miRNA in vivo to significantly limit target RNA translation in the whole heart (90).
The miRNA-based therapy to inhibit an overexpressed miRNA during diseased condition or to mimic a disease–down-regulated miRNA is schematized in Figure 3.
In spite of these encouraging premises, there is still much to learn and several concerns need to be addressed before ncRNAs can be deployed as a therapeutic option in cardiovascular conditions. Indeed, the fact that miRNA or long ncRNAs exert broad effects on multiple pathological pathways can be viewed as a major limitation with regard to both therapeutic efficacy and “off-targets” systemic effects (4-6). Therefore, further research on the enhancement of target affinity, stability and specificity are required to overcome the off-target effects and potential toxicity before ncRNA therapeutics can be used safely and effectively in the clinical setting.
Further studies are warranted to exhaustively elucidate ncRNAs changes and their underlying mechanisms in various cardiovascular pathological settings, especially regarding the cardiac function of the lncRNAs and circRNAs.
From a clinical viewpoint, ncRNAs are emerging as novel biomarkers for the diagnosis and disease progression of different cardiovascular conditions.
Nevertheless, several problems still remain unresolved mainly due to the lack of reproducibility across different studies that may hamper the transition of these circulating biomarkers from promising tools to clinical practice. Several methodological aspects related to sample collection, measure methodology and normalization seem to explain the lack of reproducibility in different published studies (91,92). Thus, technological advances are necessary to ensure fast, reliable and reproducible results for the absolute quantification of circulating ncRNA. The small sample size with reduced statistical power is another relevant problem that may have contributed to many discordant published results (91). Large-scale studies performed in a collaborative manner are also required to further validate the potential of ncRNAs as biomarkers as well as to facilitate their transfer into clinics.
In conclusion, ncRNA research is a very fascinating and challenging field that certainly will improve the knowledge of the molecular mechanisms at the basis of some cardiovascular conditions, giving the opportunity to develop new diagnostic and therapeutic approaches. However, it is a long road from a proof of concept to their widespread use as early and specific biomarkers of CVDs. It is appealing to identify specific biomarkers other than the ones conventionally used, for a precise risk stratification in patients with cardiovascular conditions. More work is needed but ncRNA are here to stay.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.11.03). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011;12:87-8. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Fiedler J, Baker AH, Dimmeler S, et al. Non-coding RNAs in vascular disease - from basic science to clinical applications: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res 2018;114:1281-86. [Crossref] [PubMed]
- Poller W, Dimmeler S, Heymans S, et al. Non coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J 2018;39:2704-16. [Crossref] [PubMed]
- Zhao G. Significance of non-coding circular RNAs and micro RNAs in the pathogenesis of cardiovascular diseases. J Med Genet 2018;55:713-20. [Crossref] [PubMed]
- Das A, Samidurai A, Salloum FN. Deciphering Non-coding RNAs in Cardiovascular Health and Disease. Front Cardiovasc Med 2018;5:73. [Crossref] [PubMed]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005;6:376-85. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Zhuo Y, Gao G, Shi JA, et al. miRNAs: biogenesis, origin and evolution, functions on virus-host interaction. Cell Physiol Biochem 2013;32:499-510. [Crossref] [PubMed]
- Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res 2012;22:1243-54. [Crossref] [PubMed]
- Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci 2008;114:699-706. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. [Crossref] [PubMed]
- Matkovich SJ, Van Booven DJ, Youker KA, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 2009;119:1263-71. [Crossref] [PubMed]
- Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J Mol Cell Cardiol 2016;94:107-21. [Crossref] [PubMed]
- Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009;119:2772-86. [Crossref] [PubMed]
- Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007;100:416-24. [Crossref] [PubMed]
- Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics 2007;31:367-73. [Crossref] [PubMed]
- Bostjancic E, Zidar N, Stajer D, et al. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology 2010;115:163-9. [Crossref] [PubMed]
- E S. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta Pharmacol Sin 2018;39:1085-99. [Crossref] [PubMed]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 2008;79:581-8. [Crossref] [PubMed]
- Schober A, Nazari-Jahantigh M, Weber C. MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis. Nat Rev Cardiol 2015;12:361-74. [Crossref] [PubMed]
- Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008;15:272-84. [Crossref] [PubMed]
- Torella D, Iaconetti C, Catalucci D, et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ Res 2011;109:880-93. [Crossref] [PubMed]
- Rayner KJ, Suárez Y, Dávalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328:1570-3. [Crossref] [PubMed]
- Moore KJ, Rayner KJ, Suarez Y, et al. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annu Rev Nutr 2011;31:49-63. [Crossref] [PubMed]
- Oury C, Servais L, Bouznad N, et al. MicroRNAs in Valvular Heart Diseases: Potential Role as Markers and Actors of Valvular and Cardiac Remodeling. Int J Mol Sci 2016;17:7. [Crossref] [PubMed]
- van der Ven CF, Wu PJ, Tibbitt MW, et al. In vitro 3D model and miRNA drug delivery to target calcific aortic valve disease. Clin Sci 2017;131:181-195. [Crossref] [PubMed]
- Vavuranakis M, Kariori M, Vrachatis D, et al. MicroRNAs in aortic disease. Curr Top Med Chem 2013;13:1559-72. [Crossref] [PubMed]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-7. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Wang Y, Hou J, He D, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet 2016;32:211-24. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77-94. [Crossref] [PubMed]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011;12:136-49. [Crossref] [PubMed]
- Klattenhoff CA, Scheuermann JC, Surface LE, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570-83. [Crossref] [PubMed]
- Liu J, Li Y, Lin B, et al. HBL1 is a human long noncoding rna that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev Cell 2017;42:333-348.e5. [Crossref] [PubMed]
- Vausort M, Wagner DR, Devaux Y. Long Noncoding RNAs in Patients with Acute Myocardial Infarction Novelty and Significance. Circ Res 2014;115:668-77. [Crossref] [PubMed]
- Zangrando J, Zhang L, Vausort M, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 2014;15:460. [Crossref] [PubMed]
- Ounzain S, Micheletti R, Beckmann T, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding rnas. Eur Heart J 2015;36:353-68a. [Crossref] [PubMed]
- Guo Y, Luo F, Liu Q, et al. Regulatory non Luo F, RNAs in acute myocardial infarction. J Cell Mol Med 2017;21:1013-23. [Crossref] [PubMed]
- Yang KC, Yamada KA, Patel AY, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014;129:1009-21. [Crossref] [PubMed]
- Greco S, Zaccagnini G, Perfetti A, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med 2016;14:183. [Crossref] [PubMed]
- Pang L, Hu J, Zhang G, et al. Dysregulated long intergenic non-coding RNA modules contribute to heart failure. Oncotarget 2016;7:59676-690. [Crossref] [PubMed]
- Wang K, Liu F, Zhou LY, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 2014;114:1377-88. [Crossref] [PubMed]
- Micheletti R, Plaisance I, Abraham BJ, et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med 2017;9:eaai9118.
- Ishii N, Ozaki K, Sato H, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-99. [Crossref] [PubMed]
- Qu X, Du Y, Shu Y, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep 2017;7:42657. [Crossref] [PubMed]
- Zhu XH, Yuan YX, Rao SL, et al. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci 2016;20:3653-60. [PubMed]
- Li Y, Wang J, Sun L, et al. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol 2018;818:508-17. [Crossref] [PubMed]
- Viereck J, Kumarswamy R, Foinquinos A, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 2016;8:326ra22 [Crossref] [PubMed]
- Han P, Li W, Lin CH, et al. A long noncoding rna protects the heart from pathological hypertrophy. Nature 2014;514:102-6. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-06. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-88. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. [Crossref] [PubMed]
- Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [PubMed]
- Geng HH, Li R, Su YM, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016;11:e0151753 [Crossref] [PubMed]
- Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342. [Crossref] [PubMed]
- Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun 2017;487:769-75. [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Khan MA, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the Titin gene. Circ Res 2016;119:996-1003. [Crossref] [PubMed]
- Lei W, Feng T, Fang X, et al. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther 2018;9:56. [Crossref] [PubMed]
- Anfossi S, Babayan A, Pantel K, et al. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol 2018;15:541-63. [Crossref] [PubMed]
- Viereck J, Thum T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ Res 2017;120:381-99. [Crossref] [PubMed]
- Navickas R, Gal D, Laucevicius A, et al. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res 2016;111:322-37. [Crossref] [PubMed]
- Cavarretta E, Frati G. MicroRNAs in Coronary Heart Disease: Ready to Enter the Clinical Arena? Biomed Res Int 2016;2016:2150763 [Crossref] [PubMed]
- Villar AV, Garcia R, Merino D, et al. Myocardial and circulating levels of microRNA-21 reflect left ventricular fibrosis in aortic stenosis patients. Int J Cardiol 2013;167:2875-81. [Crossref] [PubMed]
- García R, Villar AV, Cobo M, et al. Circulating levels of miR-133a predict the regression potential of left ventricular hypertrophy after valve replacement surgery in patients with aortic stenosis. J Am Heart Assoc 2013;2:e000211 [Crossref] [PubMed]
- Zhang Y, Sun L, Xuan L, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep 2016;6:22384. [Crossref] [PubMed]
- Yan Y, Zhang B, Liu N, et al. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int 2016;2016:8079372 [Crossref] [PubMed]
- Li M, Wang YF, Yang XC, et al. Circulating Long Noncoding RNA LIPCAR Acts as a Novel Biomarker in Patients with ST-Segment Elevation Myocardial Infarction. Med Sci Monit 2018;24:5064-5070. [Crossref] [PubMed]
- Gao L, Liu Y, Guo S, et al. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell Physiol Biochem 2017;44:1497-08. [PubMed]
- Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014;114:1569-75. [Crossref] [PubMed]
- Xuan L, Sun L, Zhang Y, et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J Cell Mol Med 2017;21:1803-1814. [Crossref] [PubMed]
- Memczak S, Papavasileiou P, Peters O, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 2015;10:e0141214 [Crossref] [PubMed]
- Vausort M, Salgado-Somoza A, Zhang L, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol 2016;68:1247-48. [Crossref] [PubMed]
- Deng YY, Zhang WP, She JQ, et al. GW27 YY, Z circular RNA related to PPARγ function as ceRNA of microRNA in human acute myocardial infarction. J Am Coll Cardiol 2016;68:C51-2. [Crossref]
- Burd CE, Jeck WR, Liu Y, et al. non4/ARF Y RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233 [Crossref] [PubMed]
- Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA Hsa_Circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918. [Crossref] [PubMed]
- Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011;478:404-07. [Crossref] [PubMed]
- Boon RA, Seeger T, Heydt S, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res 2011;109:1115-19. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Fang J, Song XW, Tian J, et al. Overexpression of microrna-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 2012;17:410-23. [Crossref] [PubMed]
- Ganesan J, Ramanujam D, Sassi Y, A, et al. Mir-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation 2013;127:2097-106. [Crossref] [PubMed]
- O'Donnell JM, Kalichira A, Bi J, et al. In vivo, cardiac-specific knockdown of a target protein, malic enzyme-1, in rat via adenoviral delivery of DNA for non-native miRNA. Curr Gene Ther 2012;12:454-62. [Crossref] [PubMed]
- de Gonzalo-Calvo D, Vea A, Bär C, et al. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem 2015;61:56-63. [Crossref] [PubMed]
Cite this article as: Andreassi MG. Non-coding RNA in cardiovascular disease: a general overview on microRNAs, long non-coding RNAs and circular RNAs. Non-coding RNA Investig 2018;2:63.


