The effects of non-coding RNAs on autophagy in regulating cardiac hypertrophy
Introduction
Cardiovascular disease is the most prevalent disease worldwide and is one of the leading causes of mortality all over the world. Cardiac hypertrophy is an essential predictor of progressive cardiovascular diseases with a poor prognosis. Initially, cardiac hypertrophy serves as a compensatory response for the biomechanical and pathophysiological stimuli, and the increase of heart size and mass are accompanied by biochemical, molecular, structural and metabolic changes to maintain cardiac contractile function (1). Nevertheless, prolonged hypertrophy may eventually progress to detrimental cardiac remodeling, leading to heart failure and even sudden death. At the cellular level, cardiac hypertrophy is characterized by enlargement of cardiomyocyte size, increased protein synthesis, reorganization of sarcomeric structure and re-expression of fetal genes (2). In the past decades, the molecular mechanisms involved in the development of cardiac hypertrophy have been investigated in multiple physiological processes, such as autophagy, modification of DNA and histones, metabolism and oxidative stress (3).
Recent genome-wide studies have shown that while only 2% of the mammalian genome encodes mRNAs, the vast majority is pervasively transcribed as non-protein-coding RNAs (ncRNAs), including miRNAs, small interfering RNAs, PIWI-interacting RNAs and long ncRNAs. ncRNAs function as transcriptional and post-transcriptional regulators and as guides of chromatin-modifying complexes, involved in normal development and pathophysiology for diseases (4). MicroRNAs (miRNAs) are small ncRNAs that are ~22 nucleotides in length, produced by RNase III proteins Drosha and Dicer. In eukaryotic cells, miRNAs function as guide molecules in RNA silencing through base pairing with its target mRNAs and subsequently triggering mRNA degradation (5). Owing to the inhibitory effect on numerous protein-coding transcripts, miRNAs are involved in almost all aspects of developmental and pathological process in eukaryotes. Identification of target mRNAs for specific miRNAs is essential for investigating physiological function of miRNAs. In the past decade, studies utilizing pleiotropic cellular and animal disease models have allowed identification of miRNAs and their specific target genes in cardiovascular development and various cardiovascular diseases (6). Inhibition of miR-27b using a specific antagomir in pressure-overload-induced heart failure attenuated cardiac hypertrophy and dysfunction through targeting peroxisome proliferator-activated receptor-γ (PPARγ), a well-known regulator of cardiac hypertrophy (7). Similarly, cardiac-specific deletion of miR-22 extenuated isoproterenol-induced cardiac hypertrophy and dysfunction through directly downregulating sirtuin 1 (SIRT1) and histone deacetylase 4 (HDAC4) (8). In addition, overexpression of miR-21-3p significantly suppressed transverse aortic constriction (TAC)-induced and angiotensin II- induced cardiac hypertrophy. Histone deacetylase-8 (HDAC8) is proved to be the direct target of miR-21-3p, and re-expression of HDAC8 diminished miR-21-3p-mediated inhibition of cardiac hypertrophy through Akt/Gsk3β pathway (9).
ncRNAs as autophagy regulators
Autophagy is an evolutionarily conserved intracellular catabolic process involving degradation of proteins and organelles through lysosomes (10). Basal autophagy is a cytoprotective mechanism in maintaining cellular homeostasis through the elimination of damaged/old organelles and proteins (11). Abnormal autophagy is associated with multiple pathological disorders. As an essential intracellular process, autophagy needs to be tightly regulated.
Recently, miRNAs have been discovered to be involved in the regulation of autophagy. The regulatory effect of miRNAs on autophagy was first revealed in 2009 and miR-30a was demonstrated to regulate cellular autophagy through targeting an important autophagy-promoting gene beclin-1. The expression of miR-30a was downregulated under nutrient depletion or rapamycin treatment. Beclin-1 was negatively regulated by miR-30a both at mRNA and protein level in tumor cells. Furthermore, suppression of beclin-1 expression using miR-30a mimic declined autophagy induced by rapamycin (12). Soon after this study, numerous miRNAs involved in autophagy have been studied in certain diseases including cancers, cardiovascular diseases and Crohn disease. Another miRNA regulating beclin-1 was miR-376b. Inhibition of endogenous miR-376b by antagonist led to an increase in autophagy related 4C cysteine peptidase (ATG4C) and beclin-1 levels (13). Furthermore, a variety of miRNAs have been reported to be involved in different stages of autophagy, including induction, vesicle nucleation, vesicle elongation and maturation (14). It was reported that miR-20a and miR-106b negatively regulated autophagy by downregulating unc-51 like autophagy activating kinase 1 (ULK1), which is associated with the initiation of autophagy (15). In a functional screening for miRNAs modulating the autophagic flux in breast cancer cells, miR-101 was identified to be a potent inhibitor of autophagy. miR-101 targets three autophagy-related genes, member RAS oncogene family (RAB5A), ATG4D and stathmin 1 (STMN1). In addition, inhibition of RAB5A attenuated basal and rapamycin-induced autophagy. Through targeting RAB5A, miR-101 probably involved in both vesicle nucleation and elongation (16). A potential regulator of the maturation step was miR-130a, which is associated with the ATG9-ATG2-ATG18 complex formation via downregulating ATG2B (17). So far accumulating studies have indicated that miRNAs play critical roles in the autophagy processes.
Apart from miRNAs, long non-coding RNAs (lncRNAs) are emerging as new regulators of autophagy. A long non-coding RNA named autophagy promoting factor (APF), was reported to modulate autophagic cell death through targeting miR-188-3p and subsequently regulating downstream target ATG7 expression (18). In vascular endothelial cells, overexpression of lncRNA FLJ11812 enhanced autophagic activity by directly downregulating the level of miR-4459, and thus releasing miR-4459 downstream target ATG13 (19).
Autophagy regulates cardiac hypertrophy
The development of cardiac hypertrophy involves alterations in the balance between protein synthesis and degradation (20). Mounting studies have focused on the relationship between cardiac hypertrophy and catabolic pathways, showing that many hypertrophic related signaling pathways are involved in the regulation of autophagy (21). It is generally accepted that basal autophagy is crucial for the maintenance of cellular homeostasis, whereas excessive or insufficient autophagy can exacerbate cardiac hypertrophy and may progress to heart failure. Cardiac-specific defect of ATG5, an autophagy related gene, leads to cardiac hypertrophy and contractile dysfunction (22). Ablation of Mcl-1, an anti-apoptotic protein of BCL-2 family, leads to impaired contractility, cardiac hypertrophy and early mortality with suppressed autophagy (23). Glycogen synthase kinase 3 alpha (Gsk3α) knockout mice exert impaired autophagy with activation of mTORC1 and develop cardiac hypertrophy and contractile dysfunction with age (24). However, in a study with opposite findings, autophagic activity peaked at 48 hours after thoracic aortic banding and remained significantly enhanced for at least 3 weeks. Furthermore, disruption of beclin-1 attenuated autophagic activity and extenuated pathological remodeling, while overexpression of beclin-1 increased cardiomyocyte autophagy and aggravated remodeling (25). In addition, inhibition of histone deacetylases (HDACs) using trichostatin A (TSA) ameliorated pathologic hypertrophy and suppressed the activation of autophagy (26). These studies demonstrate conflicting opinions about the role of autophagy in cardiac hypertrophy. Differences in severity of stress and mouse models used in studies may contribute to these discrepancies (27). Besides, the dual role of autophagy in the development of cardiac hypertrophy might be associated with the stages and severity of diseases (28).
ncRNAs regulate cardiac hypertrophy through autophagy
Pro-hypertrophic and anti-autophagic miRNAs
Several studies have demonstrated that miRNAs promoted cardiac hypertrophy in a manner of negatively modulating some known pro-autophagic regulators. Upon hypertrophic conditions, miR-212 and miR-132 were upregulated both in vivo and in vitro. Overexpression of the miRNA-212/132 family could result in pathological cardiac hypertrophy and subsequent heart failure, whereas the deletion of miRNA-212/132 attenuated the development of cardiac hypertrophy induced by pressure overload. Mechanistically, miR-212/132 diametrically targeted and negatively regulated forkhead box O3 (FoxO3), which serves as a powerful anti-hypertrophic and pro-autophagic transcription factor in cardiomyocytes, and thus led to the activation of pro-hypertrophic calcineurin/NFAT pathway in pace with the inhibition of cardiac autophagy (29).
In another study performed in cardiac-specific miR-199a transgenic mice, overexpression of miR-199a led to pathological cardiac hypertrophy and impairment of cardiac function with lower cardiomyocyte autophagy. Glycogen synthase kinase 3β (Gsk3β), a pro-autophagic and anti-hypertrophic gene mediating mTOR signaling suppression, was a direct target gene of miR-199a. Furthermore, activation of autophagy using rapamycin or overexpression of ATG5 was sufficient to rescue the pro-hypertrophic effect resulted from miR-199a overexpression (30).
Similarly, cardiac-specific overexpression of miR-221 led to cardiac dysfunction and heart failure with impaired autophagy. Evidence showed that miR-221 suppressed autophagy through diametrically targeting p27, thereby releasing the p27-mediated inhibition of CDK2 and activating the mTOR signaling pathway. Reactivation of autophagy by rapamycin treatment or knockdown of raptor reversed the pro-hypertrophic effect of miR-221 overexpression. These findings clarify that miR-221 plays a crucial role in autophagy balance and cardiac hypertrophy through modulating the p27/CDK2/mTOR signaling (31).
Anti-hypertrophic and anti-autophagic miRNAs
Contrary to the effects of pro-hypertrophic and anti-autophagic miRNAs above, several studies have got some diverse findings. miR-30a was found to be an anti-autophagic and anti-hypertrophic regulator in a study, where beclin-1 was demonstrated to be a target gene of miR-30a. Downregulation of miR-30a and upregulation of beclin-1 emerge in hypertrophic hearts induced by transverse abdominal aortic constriction accompanied by increased cardiomyocyte autophagy. Overexpression of beclin-1 led to enhanced autophagy and cardiac hypertrophy, whereas silencing of beclin-1 ameliorated cardiac hypertrophy with reduced autophagic activity. Treatment with miR-30a mimic reduced angiotensin II-induced enhancement of hypertrophic markers with lower autophagic activities, while miR-30a inhibitor promoted autophagy and thus enhanced cardiomyocyte hypertrophy. These findings reveal that miR-30a suppresses cardiac hypertrophy through inhibition of autophagy (32).
Analogously, miR-34a expression was downregulated in hypertrophic cardiomyocytes induced by angiotensin II, and ATG9A, the target gene of miR-34a, was upregulated. As an integral membrane ATG protein, ATG9A is localized in the phagophore/pre-autophagosomal structure (PAS) and plays an essential role in the autophagic process. Overexpression of ATG9A enhanced autophagic activity and cardiomyocyte hypertrophy. Mechanistically, miR-34a negatively regulated angiotensin II-induced cardiomyocyte hypertrophy by suppressing ATG9A expression and cardiomyocyte autophagy (33).
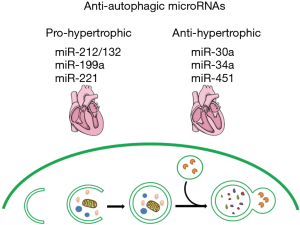
miR-451 was found to be significantly downregulated in hypertrophic cardiomyopathy (HCM) myocardium. Evidence shows that miR-451 suppressed cardiomyocyte hypertrophy by directly targeting a well-known positive autophagic regulator tuberous sclerosis complex 1 (TSC1) and inhibiting the formation of autophagosome (34). A summary of autophagy-related miRNAs regulating cardiac hypertrophy is presented in Figure 1.
Interpretation and perspective
Controversies emerge over the effects of miRNAs on cardiac hypertrophy through autophagy. According to the studies above, consisted with the current well-accepted opinion of the relationship between autophagy and cardiac hypertrophy, miR-212/132, miR-199a and miR-221 facilitate cardiac hypertrophy by inhibiting autophagic activities, with targeting several well-established hypertrophy-related signaling, such as FoxO3, Gsk3β and p27/mTOR. Conversely, miR-30a, miR-34a and miR-451 suppress autophagy and thus ameliorate cardiac hypertrophy through diametrically binding beclin-1, ATG9A and TSC1, respectively, the exact regulatory impacts of which on cardiac autophagy and hypertrophy remain pending. For instance, beclin-1, an ortholog of ATG6/vacuolar protein sorting (Vps)-30 protein, is essential for autophagosome formation through interacting with the class III-type phosphoinositide 3-kinase (class III PI3K) (35). In Zhu’s study, heterozygous disrupted beclin-1 could ameliorate the pathological remodeling induced by severe pressure overload, while the downregulated beclin-1 did not affect cardiac hypertrophy (25). However, Pan demonstrated that angiogensin II-induced cardiac hypertrophy was mediated by the upregulation of beclin-1, and miR-30a targeted decline of beclin-1 could relieve agonist-induced hypertrophy. The discrepancy between the studies might result from the different models of hypertrophy. In addition, it was found that beclin-1 could be phosphorylated at threonine 108. The translational modification enhances interaction between beclin-1 and Bcl-2/Bcl-xL and inhibits the PI3 kinase activity of the ATG14L-Beclin1-Vps34 complex, so as to retard the beclin-1 mediated autophagy (36). Therefore, it implies simply evaluating the effect of beclin-1 level on autophagy could be inadequate to some extent in certain models. Nevertheless, as an autophagy-related protein, few studies concerned about the role of ATG9A in the development of cardiac hypertrophy except the study of miR-34a, so the accurate effect of ATG9A on cardiac hypertrophy needs more evidence to determine. Studies reveal that the TSC1/2 complex is an endogenous upstream inhibitor of mTOR complex 1 (mTORC1). Deficiency of TSC1 led to severe biventricular hypertrophy and early death with activation of mTOR, while suppression of mTORC1 using rapamycin ameliorated cardiac hypertrophy induced by TSC1 knockout (36). Similarly, TSC1 transgenic mice could resist isoproterenol-induced cardiac hypertrophy with reduced mTORC1 signaling (37). However, miR-451 was proved significantly downregulated in HCM patients accompanied by enhanced TSC1. In neonatal cardiomyocytes, miR-451 could reduce cell size by targeting TSC1 and impair the formation of autophagosome. Importantly, this study did not evaluate the effects of miR-451 on TSC1 and autophagy in load- or agonists-induced cardiac hypertrophy models, thus remaining unclear that whether miR-451 could regulate cardiac hypertrophy by blocking autophagy in the context of hypertrophy.
In summary, though contradictory effects of miRNAs on cardiac hypertrophy through autophagy, it seems that the effects might attribute to the specific targets of these miRNAs, rather than the extent or severity of autophagy. Namely, the manipulation of miRNAs which targeted the well-proved hypertrophic signals like FoxO3, Gsk3β or mTOR is probably beneficial for the future treatments of cardiac hypertrophy and heart failure. Apart from miRNAs, regulation of autophagy and cardiac hypertrophy by other ncRNAs like long non-coding RNAs and circular RNAs needs to be further investigated.
Acknowledgments
Funding: This work is supported by the National Natural Science Foundation of China (No. 81770392 and 81770394).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.10.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tham YK, Bernardo BC, Ooi JY, et al. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol 2015;89:1401-38. [Crossref] [PubMed]
- Rohini A, Agrawal N, Koyani CN, et al. Molecular targets and regulators of cardiac hypertrophy. Pharmacol Res 2010;61:269-80. [Crossref] [PubMed]
- Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol 2016;97:245-62. [Crossref] [PubMed]
- Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol 2010;220:126-39. [Crossref] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [Crossref] [PubMed]
- Wang J, Liew OW, Richards AM, et al. Overview of MicroRNAs in Cardiac Hypertrophy, Fibrosis, and Apoptosis. Int J Mol Sci 2016;17: [Crossref] [PubMed]
- Wang J, Song Y, Zhang Y, et al. Cardiomyocyte overexpression of miR-27b induces cardiac hypertrophy and dysfunction in mice. Cell Res 2012;22:516-27. [Crossref] [PubMed]
- Huang ZP, Chen J, Seok HY, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res 2013;112:1234-43. [Crossref] [PubMed]
- Yan M, Chen C, Gong W, et al. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc Res 2015;105:340-52. [Crossref] [PubMed]
- Xu J, Wang Y, Tan X, et al. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 2012;8:873-82. [Crossref] [PubMed]
- Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis 2012;33:2018-25. [Crossref] [PubMed]
- Zhu H, Wu H, Liu X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009;5:816-23. [Crossref] [PubMed]
- Korkmaz G, le Sage C, Tekirdag KA, et al. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012;8:165-76. [Crossref] [PubMed]
- Sun T, Li MY, Li PF, et al. MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 2018;7. [PubMed]
- Wu H, Wang F, Hu S, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal 2012;24:2179-86. [Crossref] [PubMed]
- Frankel LB, Wen J, Lees M, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J 2011;30:4628-41. [Crossref] [PubMed]
- Kovaleva V, Mora R, Park YJ, et al. miRNA-130a Targets ATG2B and DICER1 to Inhibit Autophagy and Trigger Killing of Chronic Lymphocytic Leukemia Cells. Cancer Res 2012;72:1763-72. [Crossref] [PubMed]
- Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779. [Crossref] [PubMed]
- Ge D, Han L, Huang S, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 2014;10:957-71. [Crossref] [PubMed]
- Lavandero S, Chiong M, Rothermel BA, et al. Autophagy in cardiovascular biology. J Clin Invest 2015;125:55-64. [Crossref] [PubMed]
- Li Z, Wang J, Yang X. Functions of Autophagy in Pathological Cardiac Hypertrophy. Int J Biol Sci 2015;11:672-8. [Crossref] [PubMed]
- Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007;13:619-24. [Crossref] [PubMed]
- Thomas RL, Roberts DJ, Kubli DA, et al. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev 2013;27:1365-77. [Crossref] [PubMed]
- Zhou J, Freeman TA, Ahmad F, et al. GSK-3α is a central regulator of age-related pathologies in mice. J Clin Invest 2013;123:1821-32. [Crossref] [PubMed]
- Zhu H, Tannous P, Johnstone JL, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest 2007;117:1782-93. [Crossref] [PubMed]
- Cao DJ, Wang ZV, Battiprolu PK, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A 2011;108:4123-8. [Crossref] [PubMed]
- Nishida K, Otsu K. Autophagy during cardiac remodeling. J Mol Cell Cardiol 2016;95:11-8. [Crossref] [PubMed]
- Li L, Xu J, He L, et al. The role of autophagy in cardiac hypertrophy. Acta Biochim Biophys Sin (Shanghai) 2016;48:491-500. [Crossref] [PubMed]
- Ucar A, Gupta SK, Fiedler J, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078. [Crossref] [PubMed]
- Li Z, Song Y, Liu L, et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ 2017;24:1205-13. [Crossref] [PubMed]
- Su M, Wang J, Wang C, et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ 2015;22:986-99. [Crossref] [PubMed]
- Pan W, Zhong Y, Cheng C, et al. MiR-30-Regulated Autophagy Mediates Angiotensin II-Induced Myocardial Hypertrophy. PLoS One 2013;8:e53950 [Crossref] [PubMed]
- Huang J, Sun W, Huang H, et al. miR-34a Modulates Angiotensin II-Induced Myocardial Hypertrophy by Direct Inhibition of ATG9A Expression and Autophagic Activity. PLoS One 2014;9:e94382 [Crossref] [PubMed]
- Song L, Su M, Wang S, et al. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med 2014;18:2266-74. [Crossref] [PubMed]
- Maejima Y, Isobe M, Sadoshima J. Regulation of autophagy by Beclin 1 in the heart. J Mol Cell Cardiol 2016;95:19-25. [Crossref] [PubMed]
- Malhowski AJ, Hira H, Bashiruddin S, et al. Smooth muscle protein-22-mediated deletion of Tsc1 results in cardiac hypertrophy that is mTORC1-mediated and reversed by rapamycin. Hum Mol Genet 2011;20:1290-305. [Crossref] [PubMed]
- Zhang HM, Diaz V, Walsh ME, et al. Moderate lifelong overexpression of tuberous sclerosis complex 1 (TSC1) improves health and survival in mice. Sci Rep 2017;7:834. [Crossref] [PubMed]
Cite this article as: Liang Z, Dong Y, Liu C. The effects of non-coding RNAs on autophagy in regulating cardiac hypertrophy. Non-coding RNA Investig 2018;2:60.