Is microRNA replacement therapy promising treatment for cancer?
Since the discovery of small non-coding RNA, which regulates messenger RNA (mRNA) translation via complimentary RNA-RNA interaction, in 1993 (1), molecules consisting of highly conserved approximately 20 nucleotide sequences, currently known as microRNAs (miRNAs), have attracted great attention in the field of cancer biology. miRNAs regulate the expression of over 30% of essential genes for key biological processes, and more than half of miRNA sequences are located in cancer-associated genes or fragile sites (2). Numerous evidences have shown that miRNA expression is dysregulated in human cancer, and the dysregulated miRNAs have been shown to affect the key features of cancer cells, such as proliferation, inhibiting growth suppressors, modulating invasion and metastasis, cell survival, and angiogenesis (3). Thus, miRNAs can serve as either oncogenes or tumor suppressors (TS) and thus are classified into oncogenic miRNAs, named as oncomirs, and TS miRNAs (4). In general, the expression of TS miRNAs is diminished in cancer cells, and the impaired function of TS miRNAs contributes to carcinogenesis or cancer progression. Therefore, the restoration of TS miRNAs, also called miRNA replacement therapy, has rapidly gained interest in the field of cancer research (5).
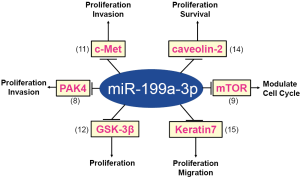
Among candidate TS miRNAs, miR-199a-3p is promising and has been gathering much attention (6,7) because its expression is known to be downregulated in several cancers including hepatocellular carcinoma (HCC) (8-10), high-grade serous ovarian carcinoma (11), renal cell cancer (12), testicular germ cell tumor (13), breast cancer (14), and bladder cancer (15). miR-199a-3p has been extensively analyzed and is known to regulate several key cancer-related genes (Figure 1). For instance, Kinose et al. showed that miR-199a-3p expression is drastically diminished in tissues of high grade serous ovarian carcinoma compared with normal ovarian epithelium, and the restoration of miR-199a-3p in cancer cells using lentivirus vector inhibited peritoneal dissemination in a xenograft model by suppressing the expression of tyrosine protein kinase Met (c-Met) (11). Dysregulated expression of miR-199a-3p has been extensively studied, especially in HCC. A comprehensive miRNA search named as miRNomes in human normal liver, hepatitis affected liver, and HCC affected liver revealed that miR-199a/b-3p, the third most enriched miRNA in normal liver, is dysregulated in HCC, and low expression of miR-199a/b-3p is significantly correlated with shortened survivals of HCC patients (8). Adenovirus-mediated in vivo gene therapy of miR-199a-3p showed its therapeutic potential in a xenograft model by inhibiting serine/threonine protein kinase P21 (RAC1) activated kinase 4 (PAK4) (8). Restoration of diminished levels of miR-199a-3p in HCC cells induced cell cycle arrest in the G1-phase, inhibited cell invasion, promoted susceptibility to hypoxia, and sensitized to doxorubicin-induced apoptosis via the inhibition of c-Met and mammalian target of rapamycin (mTOR) (9). Therefore, an efficient and selective delivery method of miR-199a-3p to HCC cells has been expected as a novel miRNA-based HCC therapy.
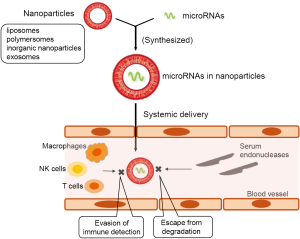
While miRNA replacement therapy has been used in preclinical trials and promising results have been reported, a few translational clinical trials for miRNA replacement therapy have failed to progress further so far (4). The main bottleneck in the systemic delivery of miRNAs is triggering of an immune response and the potential degradation of the miRNAs by endonucleases present in the extracellular space and physiological fluids. At present, viral and non-viral vectors are employed for miRNA delivery (16). The use of viral vectors is limited because of inducing immunogenicity, limited packaging capacity, and difficulty of vector production, thus, diverting the focus of researches to non-viral vectors (4). Various non-viral vector systems have been developed for DNA or miRNA delivery, including the direct injection of individual nucleotides using methods such as sonoporation, gene gun, magnetofection, and hydrodynamic delivery or the use of synthetic delivery vectors such as liposomes, polymersomes, and inorganic nanoparticles (NPs) (17). Among these methods, the entrapment of miRNAs in nanoparticulate carriers appears to be feasible for protection of miRNAs from endonuclease degradation, nonspecific interactions with proteins or non-target cells, and immune detection (Figure 2). Gaur et al. reported that chitosan nanoparticle-mediated delivery of miR-34a inhibited prostate tumor growth as well as bone metastasis in a xenograft model (18). Kamerkar et al. demonstrated that exosome-mediated delivery of siRNA, specifically targeting oncogenic KRAS in pancreatic tumor, suppressed cancer in several mouse models and extended survivals of mice (19). Exosomes are cell-derived membrane-bound vesicles of 30–150 nm in diameter, released from cells, and efficiently enter other cells (20). Unlike other synthetic nanoparticle carriers, exosomes possess transmembrane and membrane-anchored proteins which promote endocytosis, thus enhancing the delivery of their internal content such as miRNAs (21).
The development of peptide-based NPs has been achieved to deliver oligonucleotides more efficiently. Varshney et al. (22) synthesized and characterized arginine α, β-dehydrophenylalanine (RΔF) NPs as a novel carrier of miR-199a-3p to recover aberrant gene expression in HCC. Successful targeted delivery was accomplished by the conjugation of RΔF NPs (RΔF-LA NPs) with lactobionic acid (LA), a ligand for the asialoglycoprotein receptor overexpressed in HCC cell lines. RΔF-LA NPs containing miR-199a-3p (named as RΔF-LA/miR NPs) were stable in serum and against the degradation by RNase and enhanced a cellular uptake as well as an efficient delivery of miR-199a-3p to cancer cells, leading to substantial increase in miR-199a-3p levels followed by the downregulation of mTOR. In addition, intravenous injection of RΔF-LA/miR NPs into nude mice which bear HCC tumor declined more than 50% tumor growth with the inhibition of mTOR expression and increased mouse survival, suggesting that RΔF-LA/miR NPs can be an effective and promising carrier for a novel targeted miRNA replacement therapy (22).
Numerous preclinical experiments have shown that miRNAs target and suppress various oncogenic pathways, and therefore, the restoration of TS miRNAs such as miR199a-3p in cancer tissues may be a promising strategy to treat cancer. In spite of several challenges in miRNA replacement therapy which resulted in failure, clinical trials with miRNA mimics have been attempted. In 2017, van Zandwijk et al. reported a phase I TargomiR trial with miR-16 mimics to treat patients suffering from malignant pleural mesothelioma (NCT02369198) (23). They found an acceptable safety profile and early signs of activity of TargomiRs and decided to enter a phase II study. Improvement of miRNA delivery methods through the development of more specific carriers with the progress of the manipulation of epigenetic factors which modulate the function of miRNAs would further enhance the efficiency and specificity of miRNA replacement therapy for cancers.
Acknowledgments
We thank Moe Matsui for her secretarial help.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Rongrong Gao (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.09.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775-89. [Crossref] [PubMed]
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [Crossref] [PubMed]
- Hosseinahli N, Aghapour M, Duijf PHG, et al. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol 2018;233:5574-88. [Crossref] [PubMed]
- Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res 2010;70:7027-30. [Crossref] [PubMed]
- Henry JC, Park JK, Jiang J, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 2010;403:120-5. [Crossref] [PubMed]
- Minna E, Romeo P, De Cecco L, et al. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget 2014;5:2513-28. [Crossref] [PubMed]
- Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011;19:232-43. [Crossref] [PubMed]
- Fornari F, Milazzo M, Chieco P, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 2010;70:5184-93. [Crossref] [PubMed]
- Zhang LF, Lou JT, Lu MH, et al. Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. EMBO J 2015;34:2671-85. [Crossref] [PubMed]
- Kinose Y, Sawada K, Nakamura K, et al. The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget 2015;6:11342-56. [Crossref] [PubMed]
- Tsukigi M, Bilim V, Yuuki K, et al. Re-expression of miR-199a suppresses renal cancer cell proliferation and survival by targeting gsk-3beta. Cancer Lett 2012;315:189-97. [Crossref] [PubMed]
- Cheung HH, Davis AJ, Lee TL, et al. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene 2011;30:3404-15. [Crossref] [PubMed]
- Shatseva T, Lee DY, Deng Z, et al. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci 2011;124:2826-36. [Crossref] [PubMed]
- Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microrna targets based on microrna signatures in bladder cancer. Int J Cancer 2009;125:345-52. [Crossref] [PubMed]
- Bakhshandeh B, Soleimani M, Hafizi M, et al. A comparative study on nonviral genetic modifications in cord blood and bone marrow mesenchymal stem cells. Cytotechnology 2012;64:523-40. [Crossref] [PubMed]
- Yin H, Kanasty RL, Eltoukhy AA, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet 2014;15:541-55. [Crossref] [PubMed]
- Gaur S, Wen Y, Song JH, et al. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget 2015;6:29161-77. [Crossref] [PubMed]
- Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017;546:498-503. [PubMed]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. [Crossref] [PubMed]
- Vader P, Mol EA, Pasterkamp G, et al. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev 2016;106:148-56. [Crossref] [PubMed]
- Varshney A, Panda JJ, Singh AK, et al. Targeted delivery of microRNA-199a-3p using self-assembled dipeptide nanoparticles efficiently reduces hepatocellular carcinoma in mice. Hepatology 2018;67:1392-407. [Crossref] [PubMed]
- van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 2017;18:1386-96. [Crossref] [PubMed]
Cite this article as: Kobayashi M, Sawada K, Kimura T. Is microRNA replacement therapy promising treatment for cancer? Non-coding RNA Investig 2018;2:56.