Long non-coding RNAs in diabetes and diabetic cardiomyopathy
Introduction
Diabetes affects 425 million people worldwide according to the data of 2017. Diabetes is affecting more and more people nowadays because of increasing population of aging and obesity, sedentary lifestyles, and unhealthy eating habits. Diabetes can be divided into 2 types according to the pathogenesis. Type 1 diabetes (T1DM) is mainly caused by dysfunction of the islet and insulin secretion while type 2 diabetes (T2DM) is more attributed to the impaired insulin sensitivity in insulin-targeted tissues (1-3). DCM, defined as myocardial dysfunction occurring in diabetic patients without coronary artery disease, hypertension or valvular heart disease, is one of the important cardiovascular complications of diabetes, leading to heart failure and increased mortality (4). In recent years, more and more studies are focusing on the molecular mechanisms underlying diabetic cardiomyopathy (DCM).
Long non-coding RNAs (lncRNAs) are defined as RNAs longer than 200 nucleotides without protein-coding function. LncRNAs regulate gene expression via various ways. For example, they can directly act on the genomic DNA to regulate its expression. In addition, they can interact with proteins such as transcription factors and RNA-binding proteins, thus indirectly regulating gene transcription. Moreover, they can play as competing endogenous RNAs to sponge microRNA (miRNAs, miRs) and regulate the miRNA targets (5).
LncRNAs have been shown to play a role in many tissues and many diseases, regulating various physiological and pathological processes. This review aims to describe the role of lncRNAs in diabetes and DCM reported in recent 5 years. In detail, we will discuss the changes of lncRNA expression in insulin secretion, insulin resistance and DCM, as well as the underlying mechanisms.
LncRNAs regulate β-cell function and insulin secretion
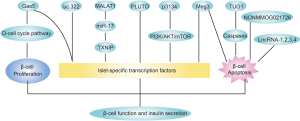
The pancreatic β-cell and its endocrine product insulin play a central role in glucose metabolism and the pathogenesis of diabetes. Although genome-wide association studies (GWASs) had identified many common genetic variants in lncRNAs associated with diabetes in the early 2010s (6-8), there were few studies to date investigating the association between lncRNAs and β-cell function until recent 5 years. Along with the growing studies about lncRNA functions in recent years, more and more researchers have focused on the role of lncRNAs in insulin secretion (Figure 1).
LncRNA TUG1 was highly expressed in the mouse pancreas compared to other tissues, implying that TUG1 is a pancreatic specific lncRNA and plays an important role in β-cell function. Downregulation of TUG1 caused β-cells apoptosis both in vivo and in vitro via caspase pathway. TUG1 expression was inhibited by high glucose and inhibition of TUG1 induces impairment of GSIS (glucose-stimulated insulin secretion) in Min6 cells. Furthermore, TUG1 inhibition in normal mice also reduced insulin secretion. Though TUG1 showed a significant association with insulin secretion, the underlying mechanism was not clarified in that study (9).
Similar to TUG1, lncRNA Meg3 expression was also dynamically regulated by glucose. Inhibition of Meg3 caused β-cell apoptosis and impaired insulin synthesis and secretion in normal mice. Suppression of Meg3 in Min6 cells decreased insulin synthesis by downregulating Pdx1 and MafA (10). Pdx1 plays an important role in pancreas development and adult β-cell function while MafA was specifically expressed in mature β-cells, which are both islet-specific transcription factors and coordinately stimulate insulin synthesis by stimulating the insulin gene promoter in response to elevated blood glucose (11). It was also observed that Meg3 was primarily localized in the nucleus and bound to EZH2 via trimethylation of H3K27 in the Rad21, Smc3, and Sin3α promoter, and in turn suppressed the expression of these transcription factors, which upregulated the expression of MafA, thus promoting the synthesis of insulin in vivo and in vitro (12). EZH2 is a methyltransferase belonging to the core component of the polycomb repressive complex-2 (PRC2), which is responsible for the trimethylation of H3K27 (13). EZH2 also interacted with Meg3 in mouse embryonic stem cells (14,15).
Besides TUG1 and Meg3, lncRNA Gas5 was also relatively high expressed in mouse pancreas and decreased in T2DM mouse models, dynamically regulated by glucose and affecting GSIG. In addition to Pdx1 and MafA, GLUT2 was also reduced in Gas5-knockdown cell line and primary islet. GLUT2 is a glucose transporter essential for activating glucose-sensitive genes and inactivation of GLUT2 leads to impaired GSIS (16). Moreover, inhibition of Gas5 could result in cell cycle G1 arrest and suppress proliferation in Min6 β-cell lines via downregulating D-cell cycle protein pathways (17).
Like the above lncRNAs, lncRNA p3134 also enhances insulin synthesis and secretion via upregulation of islet-specific transcription factors (Pdx1, MafA and GLUT2) both in vitro and in vivo. But p3134 is mainly expressed in adipose tissue and circulating exosomes secreted by islet β-cells. Additionally, TCF7L2, which was mapped to established T2D susceptibility genes, was increased by p3134 in Min6 cells and obese mice (18). Variants of TCF7L2 was correlated with impaired GSIS in humans and selective deletion of TCF7L2 in vitro induced β-cell apoptosis (19). Furthermore, PI3K/Akt/mTOR insulin signaling pathway also mediated the protection of p3134 on pancreatic islet-specific transcription factors and β-cell function in obese mice (20).
LncRNA PLUTO was restrictedly expressed in human pancreatic β-cells and PLUTO was also observed to regulate Pdx1 by affecting local 3D chromatin structure and transcription of Pdx1. PLUTO and Pdx1 were both downregulated in islets from donors with T2DMor impaired glucose tolerance, implying that PLUTO is engaged in the pathophysiology of insulin secretion (21).
LncRNA uc.322 overexpression also increased the insulin transcription factors (Pdx1 and Foxo1) and insulin secretion. LncRNA uc.322 was located in the exon region of the SOX6 gene and positively regulated SOX6 expression, which promoted the proliferation and differentiation of pancreatic islet cells (22). LncRNA MALAT1 was found increased in CSE (cigarette smoke extract)-treated Min6 cells. CSE suppressed the production of insulin by increasing MALAT1 and decreasing miR-17/TXNIP pathway (23).
On the other hand, lncRNAs associated with inflammation may also play roles in pancreatic function. LncRNAs (lncRNA-1,2,3,4) regulated by proinflammatory cytokines were increased with the development of insulitis in non-obese diabetic (NOD) mice and increased β-cell apoptosis, suggesting that they may contribute to cytokine-mediated β-cell dysfunction occurring in the initial phases of T1DM (24). Duodenum lncRNAs also participated in pancreatic secretion and inflammatory processes during the duodenal-jejunal bypass (DJB) operation, which could improve glycemic level, implying that bypass of the duodenum may initiate insulin secretion and attenuate inflammation (25).
LncRNAs in glucose metabolism and insulin resistance
Insulin resistance means the failure of target tissues to respond adequately to circulating insulin, finally leading to hyperglycemia and glucose metabolism disorders in insulin-targeted tissues. Insulin resistance in the liver cells is characterized by impaired glycogen synthesis and increased glucose production. Insulin resistance in the skeletal muscle and adipose tissue is manifested as reduced glucose uptake and consumption. Until now, most studies are regard to lncRNAs engaged in insulin resistance in the liver, especially in the liver of obese T2DM (Table 1).
Table 1
| Tissue | LncRNA | Downstream targets | Function |
|---|---|---|---|
| Liver | H19 | Hnf4a | Hepatic glucose production |
| MALAT1 | JNK/IRS-1/AKT | ROS generation in liver and pancreas | |
| nuclear SREBP-1c | Hepatic lipid accumulation | ||
| Meg3 | Foxo1 | Hepatic gluconeogenesis and glycogen synthesis | |
| Risa | Akt & Gsk3β | Hepatic autophagy | |
| LncSHGL | CaM/PI3K/Akt/mTOR/SREBP-1c | Hepatic gluconeogenic and lipogenic gene expression | |
| Gomafu | miR-139/Foxo1 | Hepatic gluconeogenesis | |
| Skeletal muscle | H19 | let-7/Insr & Lpl | Glucose regulation in muscle |
| Adipose tissue | Blnc1 | Proinflammatory cytokine signaling | Adipose tissue inflammation and fibrosis |
Liver
Excessive hepatic glucose production (HGP) contributes significantly to the hyperglycemia of T2DM. Liver-specific H19 overexpression promotes HGP, hyperglycemia, and insulin resistance, while H19 depletion enhances insulin-dependent suppression of HGP. Further genome-wide methylation and transcriptome analyses showed that H19 knockdown altered promoter methylation of Hnf4a, which is a gluconeogenic transcription factor in hepatic cells (26).
Similar to H19, lncRNA Meg3 overexpression also increased hepatic gluconeogenesis and suppressed insulin-stimulated glycogen synthesis in primary hepatocytes via increasing Foxo1 expression. Meg3 expression was enhanced by high fat through histone acetylation in hepatocytes while Meg3 interference could improve glucose tolerance and glycogen content in obese mice (27).
LncSHGL (lncRNA suppressor of hepatic gluconeogenesis and lipogenesis) and its human homologous lncRNA B4GALT1-AS1 were reduced in the livers of obese mice and patients with fatty liver disease. LncSHGL overexpression repressed gluconeogenic and lipogenic gene expression, thus improving hyperglycemia, insulin resistance, and steatosis in obese diabetic mice. Whereas lncSHGL inhibition exerted the opposite effects in normal mouse livers, promoting fasting hyperglycemia and lipid deposition. Moreover, lncSHGL recruited heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) to enhance the CaM/PI3K/Akt pathway and repress the mTOR/SREBP-1c pathway independent of calcium and insulin in hepatocytes, playing as an insulin-independent suppressor of hepatic gluconeogenesis and lipogenesis (28).
Besides the above, hepatic lncRNA Gomafu was also associated with gluconeogenesis in both obese and normal mice. Knockdown of Gomafu improved insulin sensitivity in obese mice while overexpression of it led to an increase of fasting blood glucose level in lean mice. Further mechanism research showed that in the obese mouse liver, Gomafu was upregulated by NF-κB, sponged miR-139 and then increased Foxo1 expression, finally inducing inappropriate activation of gluconeogenesis (29).
Although no evidence showed that MALAT1 was engaged in gluconeogenesis and glycogen synthesis, it was extensively studied in insulin resistance. Downregulation of MALAT1 prevented hepatic lipid accumulation and insulin resistance both in vivo and in vitro by decreasing nuclear SREBP-1c protein (30). MALAT1 ablation attenuated ROS generation in liver and pancreas, thus improving ROS-induced insulin resistance via inhibiting JNK activity and the subsequent activation of IRS-1 and phosphorylation of Akt. Meanwhile, MALAT1 ablation sensitized insulin response to fast-refeeding and hyperglycemia both in MALAT1 null mice and in isolated MALAT1 null islets, implying MALAT1 is an important therapeutic target for T2DM (31).
Altered autophagic activity is implicated in the progression of obesity to T2DM through impaired β-cell function and development of insulin resistance. A lncRNA named Risa was identified as a regulator of insulin sensitivity and autophagy. Knocking down Risa alleviated insulin resistance via enhancing hepatic autophagy in both normal and obese mice while overexpression of Risa in primary hepatocytes or C2C12 myotubes induces insulin resistance via suppressing insulin-stimulated phosphorylation of Akt and Gsk3β (32).
Skeletal muscle
H19 was downregulated in skeletal muscle of both human subjects with T2DM and insulin resistant mice as compared to healthy controls, implying that H19 is associated with insulin resistance in skeletal muscle. In vitro experiments showed that H19 depletion impaired insulin sensitivity of muscle cells via activating let-7, which in turn inhibited key metabolic genes such as Insr and Lpl. On the other hand, let-7 targeted H19 for degradation during acute hyperinsulinemia in non-diabetic muscle, illustrating a double-negative feedback loop between H19 and let-7 that contributes to glucose regulation in muscle (33).
Adipose tissue
Blnc1 is upregulated in fats from obese mice and fat-specific inactivation of Blnc1 exacerbates adipose tissue inflammation and fibrosis, leading to insulin resistance and hepatic steatosis, while overexpression of Blnc1 in adipose tissue preserves systemic metabolic homeostasis. Mechanistically, Blnc1 attenuated proinflammatory cytokine signaling and promoted fuel storage in adipocytes through its protein partner Zbtb7b (34).
LncRNA and DCM
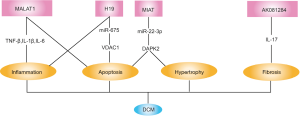
Insulin resistance in cardiomyocytes induces decreased mitochondrial glucose utilization and enhanced fatty acid uptake, resulting in triacylglycerol and cytotoxic lipid species accumulating as well as lipid metabolic gene reconstruction in the cardiomyocyte, which is recognized to precede the onset of left ventricular dysfunction of DCM (35). Role of lncRNAs in DCM has been explored in recent years and it is observed that lncRNAs affected multiple pathological processes associated with heart failure in DCM (Figure 2).
LncRNA MALAT1 was the firstly lncRNA reported to be associated with DCM. MALAT1 was upregulated in the dilated and failure diabetic heart. Moreover, MALAT1 knockdown reduced inflammatory cytokine concentration (TNF-α, IL-1β and IL-6) in the diabetic myocardium, suggesting that MALAT1 might be involved in the inflammatory process of DCM (36). Downregulation of MALAT1 was also proved to markedly reduce cardiomyocyte apoptosis in diabetic rats (37).
Similar to MALAT1, lncRNA MIAT was also upregulated in the myocardium of diabetic rats and MIAT knockdown improved cardiac structure and function in DCM. Mechanically, MIAT played as a competing endogenous RNA to sponge miR-22-3p, subsequently upregulating DAPK2 and consequently leading to hypertrophy and apoptosis of cardiomyocytes exposure to hyperglycemia (38).
On the contrary to MALAT1 and MIAT, lncRNA H19 was downregulated in the myocardium of diabetic rats while overexpression of H19 alleviated inflammation and oxidative stress in DCM and improved structure and function of the diabetic hearts. In vitro experiments further revealed that high glucose could induce primary cardiomyocyte apoptosis by regulating H19/miR-675/VDAC1 pathway (39). VDAC1 belongs to the mitochondrial porin family and is recognized as a key protein in the process of mitochondria-mediated apoptosis (40,41).
LncRNA-AK081284 was upregulated in high glucose-treated cardiac fibroblasts and knockdown of AK081284 abolished high glucose induced collagen production. Furthermore, reverse experiments indicated that the regulation of high glucose on AK081284 was mediated by IL-17 pathway (42).
Besides the above, circulating lncRNAs LIPCAR, MIAT and SENCR were identified as biomarkers of left ventricular diastolic function and remodeling in T2DM patients by RT-PCR screening (43). LncRNA Neat1 was found downregulated in diabetic Akita hearts by microarray and next generation sequencing though the underlying mechanism is unclear (44).
Conclusions
As we know, lncRNAs play an important role in gene regulation, thus engaged in the pathogenesis of various diseases. In this review, we summarized the frontier researches regarding the role of lncRNAs in diabetes and DCM, which is a severe complication of diabetes. In detail, lncRNAs are engaged in β-cell proliferation, apoptosis and insulin secretion in the islet, as well as insulin resistance in the insulin-targeted tissues and multiple pathological processes concerning DCM, implying that lncRNAs are important therapeutic targets for diabetes and DCM.
It is worth mentioning that several lncRNAs engaged in insulin secretion or insulin resistance are also engaged in DCM, in keeping with the opinion that DCM is attributed to inappropriate fuel uptake in the cardiomyocyte, prompting that lncRNAs concerning diabetes may also play roles in diseases induced by metabolism disorder. Despite, more efforts are in need to investigate the role of lncRNAs in the onset of DCM and ventricular remodeling of the diabetic heart. It is also meaningful to explore the role of lncRNAs as biomarkers for DCM diagnosis and prognosis prediction.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.09.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang L, Gao P, Zhang M, et al. Prevalence and Ethnic Pattern of Diabetes and Prediabetes in China in 2013. JAMA 2017;317:2515-23. [Crossref] [PubMed]
- Lipscombe LL. The US diabetes epidemic: tip of the iceberg. Lancet Diabetes Endocrinol 2014;2:854-5. [Crossref] [PubMed]
- Danaei G, Finucane MM, Lu Y, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet 2011;378:31-40. [Crossref] [PubMed]
- Falcão-Pires I, Leite-Moreira AF. Diabetic cardiomyopathy: understanding the molecular and cellular basis to progress in diagnosis and treatment. Heart Fail Rev 2012;17:325-44. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579-89. [Crossref] [PubMed]
- Pasmant E, Sabbagh A, Vidaud M, et al. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 2011;25:444-8. [Crossref] [PubMed]
- Kameswaran V, Bramswig NC, McKenna LB, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab 2014;19:135-45. [Crossref] [PubMed]
- Yin DD, Zhang EB, You LH, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol BIOCHEM 2015;35:1892-904. [Crossref] [PubMed]
- You L, Wang N, Yin D, et al. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J Cell Physiol 2016;231:852-62. [Crossref] [PubMed]
- Guo QS, Zhu MY, Wang L, et al. Combined transfection of the three transcriptional factors, PDX-1, NeuroD1, and MafA, causes differentiation of bone marrow mesenchymal stem cells into insulin-producing cells. Exp Diabetes Res 2012;2012:672013 [PubMed]
- Wang N, Zhu Y, Xie M, et al. Long Noncoding RNA Meg3 Regulates Mafa Expression in Mouse Beta Cells by Inactivating Rad21, Smc3 or Sin3alpha. Cell Physiol Biochem 2018;45:2031-43. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Kaneko S, Bonasio R, Saldana-Meyer R, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 2014;53:290-300. [Crossref] [PubMed]
- Mondal T, Subhash S, Vaid R, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun 2015;6:7743. [Crossref] [PubMed]
- Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia 2015;58:221-32. [Crossref] [PubMed]
- Jin F, Wang N, Zhu Y, et al. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic beta Cells. Cell Physiol Biochem 2017;43:2062-73. [Crossref] [PubMed]
- Takamoto I, Kubota N, Nakaya K, et al. TCF7L2 in mouse pancreatic beta cells plays a crucial role in glucose homeostasis by regulating beta cell mass. Diabetologia 2014;57:542-53. [Crossref] [PubMed]
- Shu L, Zien K, Gutjahr G, et al. TCF7L2 promotes beta cell regeneration in human and mouse pancreas. Diabetologia 2012;55:3296-307. [Crossref] [PubMed]
- Ruan Y, Lin N, Ma Q, et al. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet beta-Cell Function. Cell Physiol Biochem 2018;46:335-50. [Crossref] [PubMed]
- Akerman I, Tu Z, Beucher A, et al. Human Pancreatic beta Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab 2017;25:400-11. [Crossref] [PubMed]
- McDonald E, Krishnamurthy M, Goodyer CG, Wang R. The emerging role of SOX transcription factors in pancreatic endocrine cell development and function. Stem Cells Dev 2009;18:1379-88. [Crossref] [PubMed]
- Sun Q, Xu H, Xue J, et al. MALAT1 via microRNA-17 regulation of insulin transcription is involved in the dysfunction of pancreatic beta-cells induced by cigarette smoke extract. J Cell Physiol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Motterle A, Gattesco S, Caille D, et al. Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 2015;58:1827-35. [Crossref] [PubMed]
- Liang Y, Yu B, Wang Y, et al. Duodenal long noncoding RNAs are associated with glycemic control after bariatric surgery in high-fat diet-induced diabetic mice. Surg Obes Relat Dis 2017;13:1212-26. [Crossref] [PubMed]
- Zhang N, Geng T, Wang Z, et al. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Zhu X, Wu YB, Zhou J, et al. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun 2016;469:319-25. [Crossref] [PubMed]
- Wang J, Yang W, Chen Z, et al. Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis. Diabetes 2018;67:581-93. [Crossref] [PubMed]
- Yan C, Li J, Feng S, et al. Long noncoding RNA Gomafu upregulates Foxo1 expression to promote hepatic insulin resistance by sponging miR-139-5p. Cell Death Dis 2018;9:289. [Crossref] [PubMed]
- Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep 2016;6:22640. [Crossref] [PubMed]
- Chen J, Ke S, Zhong L, et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem Pharmacol 2018;152:94-103. [Crossref] [PubMed]
- Wang Y, Hu Y, Sun C, et al. Down-regulation of Risa improves insulin sensitivity by enhancing autophagy. FASEB J 2016;30:3133-45. [Crossref] [PubMed]
- Gao Y, Wu F, Zhou J, et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res 2014;42:13799-811. [Crossref] [PubMed]
- Zhao XY, Li S, DelProposto JL, et al. The long noncoding RNA Blnc1 orchestrates homeostatic adipose tissue remodeling to preserve metabolic health. Mol Metab 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Bayeva M, Sawicki KT, Ardehali H. Taking diabetes to heart--deregulation of myocardial lipid metabolism in diabetic cardiomyopathy. J Am Heart Assoc 2013;2:e000433 [Crossref] [PubMed]
- Zhang M, Gu H, Chen J, et al. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int J Cardiol 2016;202:753-5. [Crossref] [PubMed]
- Zhang M, Gu H, Xu W, et al. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol 2016;203:214-6. [Crossref] [PubMed]
- Zhou X, Zhang W, Jin M, et al. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis 2017;8:e2929 [Crossref] [PubMed]
- Li X, Wang H, Yao B, et al. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep 2016;6:36340. [Crossref] [PubMed]
- Shoshan-Barmatz V, Mizrachi D, Keinan N. Oligomerization of the mitochondrial protein VDAC1: from structure to function and cancer therapy. Prog Mol Biol Transl Sci 2013;117:303-34. [Crossref] [PubMed]
- Shoshan-Barmatz V, Golan M. Mitochondrial VDAC1: function in cell life and death and a target for cancer therapy. Curr Med Chem 2012;19:714-35. [Crossref] [PubMed]
- Zhang Y, Zhang YY, Li TT, et al. Ablation of interleukin-17 alleviated cardiac interstitial fibrosis and improved cardiac function via inhibiting long non-coding RNA-AK081284 in diabetic mice. J Mol Cell Cardiol 2018;115:64-72. [Crossref] [PubMed]
- de Gonzalo-Calvo D, Kenneweg F, Bang C, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep 2016;6:37354. [Crossref] [PubMed]
- Kesherwani V, Shahshahan HR, Mishra PK. Cardiac transcriptome profiling of diabetic Akita mice using microarray and next generation sequencing. PLoS One 2017;12:e0182828 [Crossref] [PubMed]
Cite this article as: Shen S, Chen J, Deng X, Shen L. Long non-coding RNAs in diabetes and diabetic cardiomyopathy. Non-coding RNA Investig 2018;2:54.