
Peripheral biomarkers in glioblastoma patients—is it all just HOTAIR?
Glioblastoma multiforme is the most aggressive form of brain cancer. The standard of care treatment for newly diagnosed patients with glioblastoma is tumour de-bulking by surgery, followed combination therapy consisting of radiation and temozolomide chemotherapy (1). The tumour invariably recur, often in the same area or adjacent to as the original surgical-resected tumor (2,3). Tumour recurrence or progression is currently assessed by magnetic resonance imaging (MRI), which may include spectroscopy studies and blood flow (3). However, MRI has limitations for determining recurrence or treatment response. Therefore, identifying peripheral biomarkers that can predict tumor prognosis, tumor recurrence/progression and instruct personalised treatment strategies would be an important advance for improving overall patient outcomes in glioblastoma patients and other cancers.
Although several more established types of biomarker have been explored (Figure 1A), including circulating tumour cells, growth factor and cytokines, miRNA and circulating tumour DNA (4-8), the detection of various long non-coding RNA (LncRNA) have rapidly emerged as potential candidates for optimized glioma diagnosis, prognosis and targeted therapy. LncRNAs are approximately 200 nucleotides in length and have emerged as novel gene regulators that interact with nucleic- or protein-molecules and regulate transcription or posttranscriptional processes (9-12). LncRNAs are thought to function via four potential mechanisms: (I) guiding chromatin-modifying enzymes; (II) providing a scaffold for RNA-protein complexes; (III) acting in conjunction with transcription factors and (IV) acting as molecular decoys. Further information of these four mechanisms of actions are discussed by Bhat et al. (9,13) as it is beyond the scope of this editorial.
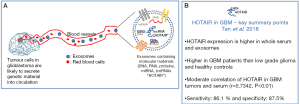
Several LncRNAs have been implicated in promoting glioblastoma progression. The study by Zhang and colleagues identified a correlation between the expression of the LncRNA HOX Transcript Antisense Intergenic RNA (HOTAIR) with tumour grade, where the highest expression of HOTAIR was observed in serum collected from glioblastoma patients compared with low grade glioma patients and healthy controls (14). HOTAIR is part of the homeobox superfamily and transcribed within the HOXC locus on chromosome 12q13. One of its known functions includes acting as a gene silencer on the HOXD locus by forming a complex with polycomb-repressive complex (PRC2) chromosome 2 (9,10,15). HOTAIR was found to be up-regulated in primary colorectal cancer biopsies and matched serum samples compared to controls (16). HOTAIR has also been identified as a strong predictor of breast cancer progression and metastasis (17). Recent evidence demonstrated that HOTAIR is also involved in the progression of other solid tumours including cervical and bladder cancer (18,19). HOTAIR expression has subsequently been shown to enhance cell cycle progression and is associated with poor overall outcome of glioblastoma patients (20,21). Likewise, the study by Pastori et al. (22) demonstrated that HOTAIR is over-expressed in human glioblastoma specimens compared to normal brain samples. Furthermore, they also showed that the knockdown of HOTAIR led to enhanced apoptosis and reduced colony formation in vitro, in addition to reduced intracranial tumour growth in vivo compared to control transfected glioblastoma cells (22).
These findings highlight the potential role of intra-tumoural HOTAIR expression in glioblastoma progression and the identification of HOTAIR LncRNA in peripheral samples such as serum, representing a likely important non-invasive biomarker for glioblastoma diagnosis and recurrence. Importantly, a follow-up article by the same group evaluated the expression levels of HOTAIR as a serum biomarker for glioblastoma diagnosis (23). HOTAIR were found to be located in exosomes and potentially not as free molecules, as it was detected in whole serum and purified exosomes, but not in serum supernatant depleted of exosomes. However, the authors did not state how many serum samples were used to harvest exosomes when displaying this data. Nevertheless, Tan et al. compared serum samples of 43 glioblastoma patients with 23 low grade glioma and 40 healthy controls and observed that HOTAIR expression was up-regulated in the glioblastoma patients (23). The sensitivity and specificity of HOTAIR expression was 86.1% and 87.5% respectively and the area under the ROC curve was 0.913 (P<0.001). The authors found a moderate correlation of HOTAIR between primary glioblastoma tumor samples and their matched serum (n=15 paired samples, r=0.7342, P<0.01) (Figure 1B). To validate serum HOTAIR as a potential biomarker, HOTAIR products was amplified and sequence mapped to its original site on chromosome 12q13 of the human genome along with the HOTAIR-205 splice variant. Serum HOTAIR expression was also monitored in one recurrent GBM patient longitudinally from before surgery to post-treatment; and found that the level of serum HOTAIR was reduced after surgery and further reduced 2-week post-surgery, suggesting its prognostic potential. The authors acknowledged that the data presented was very preliminary (n=1 recurrent), thus it is necessary to repeat this analysis with more GBM patients. More importantly, analysis of HOTAIR expression in patient serum taken at several time-points post-surgery until recurrence is detected by MRI would be critical to determine whether the re-emergence of increased HOTAIR expression correlates with the detection of tumour recurrence (or ideally found to increase prior to detection of recurrence by MRI). These longitudinal findings in a large cohort of retrospective patient serum samples would therefore determine if HOTAIR can truly be used as a peripheral biomarker for glioblastoma recurrence; and perhaps represent an improved detection method than MRI, which is currently utilized worldwide.
In summary, data from this study provides promising evidence that HOTAIR can be used as a potential diagnostic and prognostic biomarker in glioblastoma, a devastating cancer with very poor prognosis (24). It is likely that lncRNA including HOTAIR may be used as a single biomarker or a combination of biomarkers for early diagnosis and tumor monitoring. However, research in this field is still in its infancy and more experiments are required to confirm the validity of HOTAIR as a universal biomarker.
Acknowledgments
Funding: RB Luwor is a recipient of the Victorian Cancer Agency Mid-Career Research Fellowship (MCRF15017).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Quying Dai (MetroWest Medical Center, Framingham, MA, USA)
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.05.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Tully PA, Gogos AJ, Love C, et al. Reoperation for Recurrent Glioblastoma and Its Association With Survival Benefit. Neurosurgery 2016;79:678-89. [Crossref] [PubMed]
- Brandes AA, Tosoni A, Spagnolli F, et al. Disease progression or pseudoprogression after concomitant radiochemotherapy treatment: pitfalls in neurooncology. Neuro Oncol 2008;10:361-7. [Crossref] [PubMed]
- Ma C, Nguyen HPT, Luwor RB, et al. A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS One 2018;13:e0189452 [Crossref] [PubMed]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Zhao H, Shen J, Hodges TR, et al. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer 2017;16:59. [Crossref] [PubMed]
- Areeb Z, Stylli SS, Koldej R, et al. MicroRNA as potential biomarkers in Glioblastoma. J Neurooncol 2015;125:237-48. [Crossref] [PubMed]
- Gourlay J, Morokoff AP, Luwor RB, et al. The emergent role of exosomes in glioma. J Clin Neurosci 2017;35:13-23. [Crossref] [PubMed]
- Bhat SA, Ahmad SM, Mumtaz PT, et al. Long non-coding RNAs: Mechanism of action and functional utility. Non-coding RNA Research 2016;1:43-50. [Crossref]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Rutenberg-Schoenberg M, Sexton AN, Simon MD. The Properties of Long Noncoding RNAs That Regulate Chromatin. Annu Rev Genomics Hum Genet 2016;17:69-94. [Crossref] [PubMed]
- Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis 2012;48:1-8. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Liu M, Jia J, Wang X, et al. Long non-coding RNA HOTAIR promotes cervical cancer progression through regulating BCL2 via targeting miR-143-3p. Cancer Biol Ther 2018;19:391-9. [Crossref] [PubMed]
- Berrondo C, Flax J, Kucherov V, et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One 2016;11:e0147236 [Crossref] [PubMed]
- Zhou X, Ren Y, Zhang J, et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget 2015;6:8353-65. [PubMed]
- Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015;6:537-46. [PubMed]
- Pastori C, Kapranov P, Penas C, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A 2015;112:8326-31. [Crossref] [PubMed]
- Tan SK, Pastori C, Penas C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer 2018;17:74. [Crossref] [PubMed]
- Kaye AH, Morokoff A. The continuing evolution: biology and treatment of brain tumors. Neurosurgery 2014;61:100-4. [Crossref] [PubMed]
Cite this article as: Nguyen HPT, Morokoff AP, Stylli SS, Kaye AH, Luwor RB. Peripheral biomarkers in glioblastoma patients—is it all just HOTAIR? Non-coding RNA Investig 2018;2:32.


