
Heart regeneration after myocardial infarction: the role of microRNAs
Introduction
Cardiovascular diseases (CVDs) continue to be a major cause of morbidity and mortality worldwide. Despite undeniable progress in cardiology and cardiac surgery, many patients progress to heart failure after acute myocardial infarction (AMI). A major reason is that AMI induces the loss of a given amount of functional myocardial tissue, including cardiomyocytes (CMs), endothelial cells (ECs), smooth muscle cells (SMCs). All of these cell types contribute to structural, biochemical, and electrical properties of the functional heart. Unlike lower vertebrates that have a high cardiac regeneration rate, the mammal heart was for a long time considered as post-mitotic organ with no capacity to regenerate (1). The various treatments aimed to delay the onset of heart failure or to limit the consequences of CVDs, do not have the ability to replace the damaged cardiac cells and regenerate new functional myocardial tissue. The curative therapies of injured heart need both the formation of new contractile CMs and the revascularization in the injured region in order to supply sufficient oxygen and nutrition for the new-formed CMs. In this review, we discuss: (I) the heart regeneration after myocardial infarction (MI); (II) the role of microRNA (miRNA) in heart development and regeneration after MI; and (III) the therapeutic potential of modulating the expression of the miRNAs. MiRNAs are endogenous, small, highly conserved noncoding RNA molecules that have emerged as fundamental posttranscriptional regulators of gene expression.
Heart regeneration after MI
CMs renewal after MI
Most of non-mammalian animals, such as zebrafish, can regenerate the heart after myocardial injury throughout life. Multiple studies have demonstrated that zebrafish can fully restore its heart in 30 days after up to a 20% loss of ventricular tissue. This regenerative process is primarily driven by preexisting CMs proliferation (2). In mammals, CMs undergo extensive proliferation during embryogenesis and fetal development. Shortly after birth, many CMs withdraw from the proliferative cell cycle (3). Beginning at postnatal day 5 in rodents, the majority of CMs undergo a final round of DNA replication and karyokinesis in the absence of cytokinesis, which results in binucleation of approximately 90% of CMs by the second week of postnatal life (4). Binucleated CMs are thought to be incapable of division, as polyploidy generally indicates terminal differentiation. Now many studies demonstrated that new CMs are generated in human hearts during physiological ageing and after heart injuries (5-7)
The source of new CMs has been attributed to: (I) division of existing CMs (8,9); (II) progenitors residing with in the heart (10); (III) exogenous niches, such as bone marrow (11).
Studies in mice indicate that mammals retain regenerative ability after birth for only a short period (<7 days). During this period, hearts can regenerate after amputation of the ventricular apex (12) and after ischemic MI (13) through proliferation of preexisting CMs. Adult mammalian CMs can slowly self-renew at a rate of 1% of total CMs per year, declining with age. This renewal results in the exchange of ~50% of adult CMs in a human life span (5). Therefore, the induction of CMs proliferation might contribute to therapeutic cardiac regeneration.
Recent studies suggest that adult CMs can reenter the cell cycle and proliferate in response to certain mitogens such as fibroblast growth factor (FGF) (14), periostin (15), neuregulin (16). It appears that this proliferative potential is restricted to a small proportion of mononucleated CMs. The stimulation of CM proliferation as a new therapeutic strategy to induce cardiac regeneration seems still to be a challenge.
Another issue for new-formed CMs after injury is the differentiation of endogenous precursor cells. A pool of tissue-specific resident cardiac stem cells, known as cardiac progenitor cells (CPCs), was identified in the adult human myocardium. The characterization of these cells remains a difficult task, due to the lack of a highly specific marker. The c-kit, stem cell antigen-1 (Sca-1) and islet-1 (nuclear transcription factor) have been proposed (17). The participation of the endogenous CPCs to heart regeneration in physiological conditions is controversial (7,18). After injury, CPCs can be activated and may differentiae into new CMs with different stimuli, such as FGF-2 (basic FGF) (19), thymosin β (20), prostaglandin E2 (21), human stem cell factor (22), stromal-cell derived factor 1 (SDF1) (23) or brain natriuretic peptide (BNP) (10,24). These results suggest the use of CPCs might be another therapeutically issue for AMI.
Revascularization after MI
Functional regeneration of cardiac tissue, which has inherently high metabolic activity, requires CMs to have a sufficient blood supply. Therefore, the growth of new blood vessels, termed angiogenesis, is essential for the process. The precise mechanisms of revascularization following injury are not well defined.
Results from non-cardiac injury models (for example, using zebrafish fin regeneration) suggest that neovascularization during regeneration is dependent on classical angiogenic signaling mechanisms that involve various endogenous factors such as vascular endothelial growth factor receptor 2 (VEGFR2) (25), hypoxia-inducible factor 1 (HIF1-α) (26) and SDF1α (27). In rodents, activation of HIF1-α pathway improved cardiac function after MI and microvascular density in the peri-infarct area after MI (28-31). Numerous factors are expressed in the ischemic heart where they promote angiogenesis, such as VEGF, FGF. Preclinical studies showed exciting results based on protein or gene transfer delivery of these growth factors after MI, but so far, clinical trials showed mixed results in patients with AMI (32).
There is evidence to support both local proliferation of ECs and a contribution from a remote stem cell source after MI. The program of coronary vascular formation is directed by epicardium, which acts as a source of both trophic factors and progenitor cells (33). Epicardium-derived cells (EPDCs) invade the underlying myocardium and undergo epithelial-to-mesenchymal transition (EMT), which rise pericytes, SMCs and fibroblasts. The precise contribution of EPDCs to the ECs continues to be debated (34). The epicardium is quiescent in the adult heart but is reactivated and expands in response to injury (35).
In mice, reactivation of EPDC following MI demonstrates a capacity for tri-lineage differentiation into the major cardiovascular cell types (CM, SMC, and EC) (20,36,37). Studies showed that the stimulation of Tβ4 (peptide thymosin beta 4) or with modified RNA encoding VEGF-A supported neovascularization after MI through regulation of EPDC activation and subsequent differentiation into SMCs and ECs (37,38).
However, the main origin of ECs of neovessels in the injured heart was not clear. A study in 2014 that included fate mapping with Col1a2-CreER transgenic mice to label the fibroblast reported that after MI, ~35% of fibroblast-derived cells expressed the EC marker vascular endothelial (VE)-cadherin (39). A recent study used six different mouse lines expressing Cre driven by fibroblast marker, including the same Col1a2-CreER transgenic mice to trace the fate of fibroblasts after MI. They demonstrated that no fibroblasts gave rise to ECs after MI. ECs of neovessels in the injured heart derive from pre-existing EC proliferation (40). To develop novel revascularization strategies in ischemic heart disease and identify the source of neovascular ECs are important steps. More studies are needed to clarify these questions.
These data point to the possibility of a novel therapeutic strategy: stimulation of the regenerative capacity of the human heart. Cardiac regeneration research is evaluating two major avenues: (I) exogenous cell transplantation and (II) stimulation of endogenous regenerative processes. The therapeutic application of various types of cells, including progenitor cells derived from adult heart, bone marrow, or adipose tissue, has been shown to have cardioprotective effects in experimental models of AMI and in some clinical trials [reviewed in (41)]. However, the establishment of successful cell therapies is challenging owing to poor homing and survival of the implanted cells, and a limited cardiac differentiation capacity of most types of adult progenitor cells. Therefore, a therapeutic ideal- relative to cell transplantation-would be to stimulate the resident source by different agents. In this field, the critical role of miRNAs is demonstrated. Recent studies have given evidences that miRNAs play an important role in cardiac development and reparation after injury.
miRNA
Recently, a novel batch of endogenous small non-coding regulatory RNAs: miRNAs have received attention in the development of cancer and regeneration of tissue, miRNAs bind complementary sequences in target mRNAs, resulting in selective degradation or selective inhibition of their translation. miRNA is a class of natural, endogenously expressed single-stranded, small (~22 nucleotide) noncoding RNAs. The transcription of miRNAs depends on their location within the genome. MiRNAs can be located in introns of coding genes or noncoding genes or in exons. Then the miRNAs transcription depends on the host gene.
miRNA biogenesis had been detailed in review (42). Briefly, the process of miRNAs biogenesis begins when the hairpin primary miRNA (pri-miRNA) is transcribed. The maturation of miRNAs is mediated by the two RNase III endonuclease Dicer and Drosha. In the first step, the microprocessor complex composed of Drosha and DGCR8 mediates the nuclear processing of the pri-miRNA into stem-loop precursors of approximately 60–70 nucleotides (pre-miRNA). The nuclear export of the precursor is subsequently mediated by exportin-5 in a Ran-GTP dependent manner. In the second step, Dicer cleaves the pre-miRNA in the cytoplasm into the mature ~22 nucleotide miRNA, which incorporates as single-stranded RNA into the RNA-induced silencing complex (RISC). This complex directs the miRNA to the target mRNA, which leads either to translational repression or degradation of the target mRNA.
miRNAs are important modulators of all cellular pathways, especially cell fate determination, differentiation, proliferation, programmed cell death, and other processes during embryogenesis and in adult life.
Interestingly, miRNAs have recently been discovered in extracellular vesicles, in particular exosomes. Exosomes are endogenous nanovesicles of 30–100 nm in diameter. They are secreted by multiple cell types, including cardiac cell types (CMs, ECs, fibroblasts and resident stem cells), into extracellular space after fusion with the plasma membrane. The results of a number of studies suggest that exosomes derived from cardiac cells are responsible for cell-to-cell communication in the heart under both physiological and in pathological conditions. It has been shown that exosomal miRNAs can target mRNAs in recipient cells. This recent concept adds a new level of complexity to the role of miRNAs in CVDs (43).
miRNA in cardiac development
The critical role of miRNAs in cardiac development was first shown through loss of function mutations in the miRNA-processing enzyme, such as Dicer or DGCR8. A knockout of Dicer precluded the processing of pre-miRNAs into their mature form in cardiac progenitors and led to defects in heart development as well as embryonic lethality (44). Mice lacking DGCR8 in muscle tissue die prematurely with signs of heart failure and dilated cardiomyopathy (45).
MiR-1 and miR-133 families
Muscle-specific miRNAs, especially the miR-1 family and the miR-133 family, play important roles in muscle growth and differentiation. The miR-1 family and the miR-133 family express in the developing heart, and increased expression was found in neonatal mouse hearts (46). Myocyte enhancer factor 2 (MEF2) and serum response factor (SRF) cooperatively regulate the expression of two bicistronic miRNA clusters encoding miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 in cardiac and skeletal muscle (47,48). Mice lacking either miR-133a-1 or miR-133a-2 are normal, whereas deletion of both miRNAs causes lethal ventricular-septal defects (VSDs) in approximately half of double-mutant embryos or neonates; miR-133a double-mutant mice that survive to adulthood succumb to dilated cardiomyopathy and heart failure (49). Mice lacking miR-1-2 have a spectrum of abnormalities, including VSDs in a subset that suffer early lethality, cardiac rhythm disturbances in those that survive, and a striking myocyte cell-cycle abnormality that leads to hyperplasia of the heart with nuclear division persisting postnatally (44). Many genes involved during cardiogenesis were upregulated in the miR-1-2 mutants mice, such as Hrt2/Hey2 and Hand1.
MiR-15 family
In mammals, enlargement of the heart during embryonic development is primarily dependent on the increase in CM number through CM proliferation. The miR-15 family, including miR-15a, miR-15b, miR-16, miR-195, and miR-497, targets cell-cycle genes such as checkpoint kinase 1 (Chk1), who is involved in many functions during DNA repair and mitosis. Inhibition of miR-15 family was associated with the de-repression of a number of cell cycle genes in the heart and a persistence of CM mitosis beyond the normal development window of cell cycle arrest (13). Premature overexpression of miR-195 in the embryonic heart was associated with ventricular hypoplasia and ventricular septal defects in β-myosin heavy chain-miR-195 transgenic mice (13).
MyomiRs group
A number of myosin genes coexpress miRNAs, named myomiRs (miR-208a, miR-208b, and miR-499) as introns with vital purposes in cell differentiation, including the direction of cardiac myosin gene expression. Mhy6 (α-myosin heavy chain) coexpresses miR-208a, Mhy7 (β-myosin heavy chain) coexpresses miR-208b, and Mhy7b (myosin heavy chain 7b) coexpresses miR-499. MiR-208a was shown to be required for cardiac growth and expression of the β-myosin heavy chain protein, the primary contractile protein of the heart (50). Both miR-208a and miR-208b, members of the miR-208 family, target Thrap1 (thyroid hormone receptor-associated protein 1) and myostatin, important negative regulators of muscle growth. MiR-208a and miR-208b are differentially expressed during heart development parallel to the expression of their respective host genes, Mhy6 and Myh7 (50).
MiR-138 and miR-143 families
Organ patterning during embryonic development requires precise temporal and spatial regulation of protein activity. Some miRNAs inhibit protein expression and are essential for normal cardiac morphogenesis. The miR-138 family is required to establish appropriate chamber-specific gene expression patterns. They repress chondroitin sulfate proteoglycan core protein 2 (cspg2) and atrio-ventricular canal-specific transcripts in the developing ventricle via regulation of a network of development signals. This contributes to CM maturation and helps to establish the distinct identity of cardiac structures (51).
In zebrafish, miR-143 is required for chamber morphogenesis through direct repression of add3 (adducin3), which encodes an F-actin capping protein. Knockdown of miR-143 or disruption of the miR-143-add3 interaction inhibits ventricular CM F-actin remodeling, which blocks their normal growth and elongation and leads to ventricular collapse and decreased contractility (52). In addition, the expression of zebrafish miR-143 is dependent on heartbeat. Knocking-down miR-143 results in de-repression of retinoic acid signaling, and produces abnormalities in the outflow tracts and ventricles (53).
MiR-218
A highly conserved miRNA, miR-218, is encoded intronically in slit2 and slit3 and negatively regulates Robo1 and Robo2. Knockdown experiments indicate that Slit2, Robo1 and miR-218 are required for the formation of the linear heart tube in zebrafish (54). Besides, the miR-218 family is part of a regulatory circuit through tbx5, a transcription factor that mediates vertebrate cardiac development, which controls heart morphogenesis. Tbs5 overexpression affects heart development in humans and mice, resulting in heart-looping defects and chamber abnormalities (55,56)
Other miRNAs
Some other miRNAs, such as miR-126, were enriched in ECs derived from mouse embryonic stem (ES) cells and in developing mouse embryos. Knockdown of miR-126 in zebrafish and in mice resulted in loss of vascular integrity and hemorrhage during embryonic development (57,58). MiR-126 functioned in part by directly repressing negative regulators of the VEGF pathway, including the SPRED1 (Sprouty-related protein) and PIK3R2/p85-β (phosphoinositol-3 kinase regulatory subunit 2).
These articles illustrate the vital, yet not fully understood, role of miRNAs in cardiac development (Figure 1).
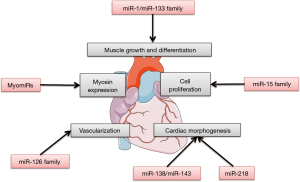
miRNA after MI
The participation of miRNA in cardiac development suggested a role of miRNAs on heart regeneration, because cardiac development is a period of high CM proliferation and vascularization. Several miRNAs have been shown to control processes that contribute to the cardiac reparation after MI (Figure 2).
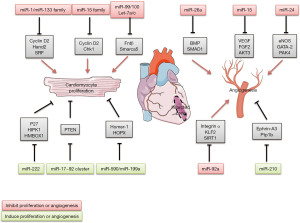
miRNA in cardiomyocyte proliferation after MI
Role of miR-15 family
Studies showed some miRNAs could modulate cell cycle and play important roles in cell reentry and CMs proliferation. MiRNAs expression profiling of hearts of neonatal mice in this period showed that the miR-15 family, plays important roles during heart development, was expressed at low levels just after birth, and upregulated at the time when CMs lost their capacity to proliferate. Knockdown of the miR-15 family in neonatal mice with locked nucleic acid (LNA)-modified anti-miRNAs was associated with an increased number of mitotic CMs (13). Although these data imply that postnatal inhibition of the miR-15 family increases CM mitotic entry and progression. Few CMs staining positive for Aurora B kinase at postnatal 12 days, suggesting that loss of miR-15 family function was not sufficient to drive complete mitotic progression through cytokinesis. Inhibition of the miR-15 family from an early postnatal age until adulthood increases myocyte proliferation in the adult heart and improves left ventricular systolic function after adult MI (9).
Role of miR-99/100 and Let-7a/c
As we previously reported, adult zebrafish heart regeneration occurs by the spontaneous dedifferentiation of resident CMs as a response to injury (2). Dedifferentiated CMs were shown to re-enter the cell cycle and accounted for all newly formed cells repairing the heart. MiR-99/100 and Let-7a/c and their protein targets fntβ (beta subunit of farnesyl-transferase) and smarca5 (SWI/SNF-related matrix associated actin-dependent regulator of chromatin subfamily a, member 5) were shown to be critical regulators of CM dedifferentiation and heart regeneration in zebrafish. Proliferation and myocardial regeneration were suggested to be mediated through their targets fntβ and smarca5.
The expression of miR-99/100 and Let-7a/c was low during zebrafish heart development and was downregulated after amputation of ventricular apex, concomitantly with high levels of fntβ and smarca5 (59). However, adult mice heart, in contrast to zebrafish, failed to regenerate their heart due to a failure in downregulating these two mRNAs after MI. Overexpression of either protein, fntβ or smarca5, in primary murine CMs in vitro resulted CM dedifferentiation and proliferation. Intracardiac delivery of an adenoviral vector encoding anti-miR-99/100 and anti-Let-7a/c significantly improved heart function 15 days after MI (59).
Role of miR-1 and miR-133 families
MiR-1 family and miR-133 family are also critical mediators of cell proliferation in cardiac muscle. In zebrafish, induced transgenic elevation of miR-133 levels after injury inhibited myocardial regeneration, while transgenic miR-133 depletion enhanced CM proliferation (60). Transgenic overexpression of miR-1 or miR-133a in the developing heart leads to a decreased pool of proliferating ventricular CMs and thin-walled ventricles (47,49). MiR-1 inhibits CM proliferation by repressing expression of the Hand2 (heart and neural crest derivatives-expressed protein 2) (47). The target of miR-133 was attributed to SRF and cyclin D2 (cell-cycle regulators G1/S-specific cyclin-D2) (49). Both these targets were involved in the cell cycle control. A recent study showed overexpression of cyclin-dependent kinase 1 (CDK1), CDK4, cyclin B1, and cyclin D1 efficiently induced cell division in post-mitotic mouse, rat and human CMs (61).
Role of miR-222
MiR-222 was shown to regulate cell proliferation and differentiation in vascular SMCs and some cancers (62,63) through in part targeting the cell-cycle inhibitor p27 (cyclin-dependent kinase inhibitor). In heart, miR-222 is necessary for exercise-induced cardiac proliferation due to its role in inhibiting p27, homeodomain-interacting protein kinase 1 (HIPK1) and homeobox containing 1 (HMBOX1) (64). Mice with inducible CMs miR-222 expression were resistant to adverse cardiac remodeling and dysfunction after ischemic injury with an increase of proliferating CMs (64).
Roles of miR-17~92 cluster
MiR-17~92 cluster, consisting of miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a, has been shown to be involve in the cardiac development. Mice deficient for miR-17~92 died shortly after birth lung hypoplasia and a ventricular septal defect. MiR-17~92 cluster has also been shown to be important for CM proliferation (65). Overexpression of miR-17~92 cluster induced CM proliferation in embryonic, neonatal and adult hearts. Moreover, overexpression of miR-17~92 cluster protected the heart from MI-induced injury in mice (66,67). PTEN (phosphatase and tensin homolog), a negative regulator of survival and proliferation, is one of the miR-17~92 cluster functional targets and is downregulated by miR-19a and miR-19b (66). Similarly, overexpression of the miR-302~367 cluster, which is a paralogs of miR-17~92 cluster (65), is sufficient to improve regeneration following adult MI; which acts via repression of the Hippo pathway (68).
Role of other miRNAs
In a high throughput overexpression approach in which 875 miRNAs were tested, at least 40 miRNAs strongly increased both DNA synthesis and cytokinesis in neonatal mouse and rat CMs. Two miRNAs in particular, miR-590 and miR-199a, were shown to promote cell-cycle re-entry of adult CMs ex vivo and to promote CM proliferation in both neonatal and adults rat CMs (69). Both, these miRNAs inhibit the expression of many negative regulators of the cell cycle, Homer protein homologue 1 (Homer1) and homeodomain-only protein homeobox (HOPX) (70). Injection of adeno-associated virus expressing miR-590 and miR-199a in the peri-infarcted area induced proliferation of CM and stimulated cardiac regeneration after MI in adult mice (69).
In a recent study, the authors compared miRNA profiles in CM between postnatal day 0 (P0) and day 10 (P10) using miRNA arrays, and found that 21 miRNAs were upregulated at P10, whereas 11 were downregulated. Among them, miR-31a-5p was identified as being able to promote CM proliferation. Neonatal rats injected with miR-31a-5p antagomir at day 0 for 3 days exhibited reduced expression of CM proliferation markers. RhoBTB1 was identified as a target gene of miR-31a-5p (71).
miRNA in revascularization after MI
Several miRNAs have been described to affect angiogenesis after MI. Initial studies of the miRNAs biogenesis enzymes Drosha and Dicer evidenced a crucial need for a balanced miRNAs expression to sustain EC integrity and angiogenic properties (72-74).
Importance of miR-17~92 cluster
The miR-17~92 cluster is also highly expressed in human ECs. As we reported this cluster consists of miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a. miR-92a and controls angiogenesis and neovascularization after AMI (75). MiR-92a was shown to inhibit endothelial-cell migration and sprouting in cell-culture studies. New pharmacological area is the use of antagomiRs. Antagomirs known as anti-miRs are a class of chemically engineered oligonucleotides that prevent other molecules from binding to a desired site on an mRNA molecule. In vivo, pharmacological inhibition of miR-92a using antagomiRs increased capillary density and improved heart function after AMI in mice (75,76). In a large-animal study, catheter-based delivery of anti-miR-92a reduced infarct size associated with a significant upregulation of capillary density (77). The targets by which miR-92a elicits its effects include integrin α5, which prevents endothelial-cell apoptosis and is important for vessel maturation (75). In addition, several vasculoprotective genes are repressed by miR-92a, such as KLF2 and SIRT1 (78).
Importance of miR-23/24/27 cluster: role of miR-24
MiR-24 belongs to miR-23/24/27 cluster, who is involved in the regulation of angiogenesis during vascular disorders and ischemic heart disease (79). MiR-24 is enriched in cardiac ECs and considerably upregulated after cardiac ischemia. Inhibition of miR-24 improved neovascularization after AMI in mice. AntagomiRs directed against miR-24 or local adenovirus-mediated miR-24 decoy delivery improved the recovery after AMI via actions on endothelial nitric oxide synthase (eNOS), GATA-2 (endothelial transcription factor), and PAK4 (serine/threonine-protein kinase) (80,81).
Importance of miR-26a
MiR-26a is commonly dysregulated in diverse cancers and CVDs, involving in various biological processes, such as proliferation, migration, invasion, angiogenesis, and metabolism by targeting multiple mRNAs. MiR-26a is also increased after AMI and inhibits angiogenesis in vitro and in vivo. Ectopic expression of miR-26a markedly induced EC cycle arrest and inhibited EC migration, sprouting angiogenesis, whereas blockade of miR-26a had the opposite effects. Overexpression inhibited formation of the caudal vein plexus in zebrafish, and exercise-induced angiogenesis was blocked in mice. Administration of a LNA-based anti-miR induced robust angiogenesis; reduced myocardial infarct size, and improve heart function. Mechanistic studies demonstrated that miR-26a inhibits the bone morphogenic protein (BMP)/SMAD1 signaling pathway in ECs (82).
Importance of miR-15 family
The miR-15 family not only regulated cell cycle genes during heart development and reduced CM proliferation, but also inhibited angiogenesis, via targeting of the well-characterized proangiogenic and endothelial survival factors: VEGF-A, FGF2 and Akt-3 (RAC-γ serine/threonine-protein kinase). On the basis of these detrimental effects on CM proliferation and vascularization, inhibition of miR-15 might be an attractive strategy to improve recovery after ischaemia preventing postischaemic heart failure (83,84).
Importance of miR-210
Recently, several studies have indicated the possibility role of miRNAs, especially miR-210, in the physiopathology of CVDs; miR-210 inducing effects on CMs and the vasculature. This miRNA is induced by hypoxia (85) and upregulated after AMI (86). In vitro, miR-210 overexpression reduced cell death. In vivo, overexpression by intramyocardial injection of minicircle vectors carrying the miR-210 precursor improved cardiac function and angiogenesis after AMI in mice (86). Minicircle DNA vectors consist of a circular expression cassette devoid of the bacterial plasmid DNA backbone; they provide several advantages including sustained transgene expression in quiescent cells/tissues. The potential targets for miR-210 are Efan3 and Ptp1b, involved in inhibition of angiogenesis and induction of apoptosis, respectively.
Conclusions and prospects
In conclusion, the end goal of complete regeneration of the infarcted heart included CM renewal, neovascularization and scar reduction. The emerging biology of endogenous regeneration is exposing a multiplicity of therapeutic targets, such as small-molecule stimulator, miRNAs or cell transplantation. Numerous roles for miRNAs in cardiac development and cardiac regeneration after MI have been identified. Several miRNAs might be attractive candidates or targets to improve recovery after MI. The inhibition or overexpression of several miRNAs after MI was shown to limit tissue injury, stimulate CM proliferation and improve neovascularization.
With advances in our understanding, new opportunities exist to manipulate miRNAs for tissue regenerative therapies in the future. However, many challenges are posed by the complexities of miRNA signaling networks and the precise spatiotemporal resolution in various diseases. For example, each miRNA targets many potential mRNAs. Identification of those mRNAs that are relevant to cardiac regeneration, especially to CM proliferation or angiogenesis, is a challenge. Further studies designed to ameliorate current therapeutic methodologies are required to develop new therapies.
Several therapeutic approaches that interfere with the molecular pathways that are involved in CVDs have shown the potential to delay and reverse cardiac dysfunction (87). In particular, modulation of cardiac metabolism, gene expression, pharmacological therapy, and miRNAs represents encouraging strategies. With the current advancements of miRNAs functions based on pathophysiological context and cell type in cardiovascular disease, we foresee new therapeutic applications for the treatment of MI in the relatively near future.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.04.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ausoni S, Sartore S. From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J Cell Biol 2009;184:357-64. [Crossref] [PubMed]
- Jopling C, Sleep E, Raya M, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606-9. [Crossref] [PubMed]
- Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res 2002;90:1044-54. [Crossref] [PubMed]
- Walsh S, Ponten A, Fleischmann BK, et al. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res 2010;86:365-73. [Crossref] [PubMed]
- Kajstura J, Gurusamy N, Ogorek B, et al. Myocyte turnover in the aging human heart. Circ Res 2010;107:1374-86. [Crossref] [PubMed]
- Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98-102. [Crossref] [PubMed]
- Hsieh PC, Segers VF, Davis ME, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 2007;13:970-4. [Crossref] [PubMed]
- Kikuchi K, Holdway JE, Werdich AA, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601-5. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A 2013;110:187-92. [Crossref] [PubMed]
- Rignault-Clerc S, Bielmann C, Liaudet L, et al. Natriuretic Peptide Receptor B modulates the proliferation of the cardiac cells expressing the Stem Cell Antigen-1. Sci Rep 2017;7:41936. [Crossref] [PubMed]
- Leri A, Rota M, Pasqualini FS, et al. Origin of cardiomyocytes in the adult heart. Circ Res 2015;116:150-66. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078-80. [Crossref] [PubMed]
- Porrello ER, Johnson BA, Aurora AB, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 2011;109:670-9. [Crossref] [PubMed]
- Engel FB, Hsieh PC, Lee RT, et al. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A 2006;103:15546-51. [Crossref] [PubMed]
- Kuhn B, del Monte F, Hajjar RJ, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med 2007;13:962-9. [Crossref] [PubMed]
- Bersell K, Arab S, Haring B, et al. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 2009;138:257-70. [Crossref] [PubMed]
- Bollini S, Smart N, Riley PR. Resident cardiac progenitor cells: at the heart of regeneration. J Mol Cell Cardiol 2011;50:296-303. [Crossref] [PubMed]
- Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013;493:433-6. [Crossref] [PubMed]
- Rosenblatt-Velin N, Lepore MG, Cartoni C, et al. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest 2005;115:1724-33. [Crossref] [PubMed]
- Smart N, Bollini S, Dube KN, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011;474:640-4. [Crossref] [PubMed]
- Hsueh YC, Wu JM, Yu CK, et al. Prostaglandin E(2) promotes post-infarction cardiomyocyte replenishment by endogenous stem cells. EMBO Mol Med 2014;6:496-503. [PubMed]
- Xiang FL, Liu Y, Lu X, et al. Cardiac-specific overexpression of human stem cell factor promotes epicardial activation and arteriogenesis after myocardial infarction. Circ Heart Fail 2014;7:831-42. [Crossref] [PubMed]
- Malliaras K, Ibrahim A, Tseliou E, et al. Stimulation of endogenous cardioblasts by exogenous cell therapy after myocardial infarction. EMBO Mol Med 2014;6:760-77. [PubMed]
- Bielmann C, Rignault-Clerc S, Liaudet L, et al. Brain natriuretic peptide is able to stimulate cardiac progenitor cell proliferation and differentiation in murine hearts after birth. Basic Res Cardiol 2015;110:455. [Crossref] [PubMed]
- Bayliss PE, Bellavance KL, Whitehead GG, et al. Chemical modulation of receptor signaling inhibits regenerative angiogenesis in adult zebrafish. Nat Chem Biol 2006;2:265-73. [Crossref] [PubMed]
- Eyries M, Siegfried G, Ciumas M, et al. Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ Res 2008;103:432-40. [Crossref] [PubMed]
- Xu C, Hasan SS, Schmidt I, et al. Arteries are formed by vein-derived endothelial tip cells. Nat Commun 2014;5:5758. [Crossref] [PubMed]
- Bao W, Qin P, Needle S, et al. Chronic inhibition of hypoxia-inducible factor prolyl 4-hydroxylase improves ventricular performance, remodeling, and vascularity after myocardial infarction in the rat. J Cardiovasc Pharmacol 2010;56:147-55. [Crossref] [PubMed]
- Jurgensen JS, Rosenberger C, Wiesener MS, et al. Persistent induction of HIF-1alpha and -2alpha in cardiomyocytes and stromal cells of ischemic myocardium. FASEB J 2004;18:1415-7. [Crossref] [PubMed]
- Nakada Y, Canseco DC, Thet S, et al. Hypoxia induces heart regeneration in adult mice. Nature 2017;541:222-7. [Crossref] [PubMed]
- Kido M, Du L, Sullivan CC, et al. Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J Am Coll Cardiol 2005;46:2116-24. [Crossref] [PubMed]
- Cochain C, Channon KM, Silvestre JS. Angiogenesis in the infarcted myocardium. Antioxid Redox Signal 2013;18:1100-13. [Crossref] [PubMed]
- Masters M, Riley PR. The epicardium signals the way towards heart regeneration. Stem Cell Res 2014;13:683-92. [Crossref] [PubMed]
- Tian X, Pu WT, Zhou B. Cellular origin and developmental program of coronary angiogenesis. Circ Res 2015;116:515-30. [Crossref] [PubMed]
- Zhou B, Pu WT. Epicardial epithelial-to-mesenchymal transition in injured heart. J Cell Mol Med 2011;15:2781-3. [Crossref] [PubMed]
- Smart N, Risebro CA, Melville AA, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 2007;445:177-82. [Crossref] [PubMed]
- Smart N, Risebro CA, Clark JE, et al. Thymosin beta4 facilitates epicardial neovascularization of the injured adult heart. Ann N Y Acad Sci 2010;1194:97-104. [Crossref] [PubMed]
- Zangi L, Lui KO, von Gise A, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 2013;31:898-907. [Crossref] [PubMed]
- Ubil E, Duan J, Pillai IC, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 2014;514:585-90. [Crossref] [PubMed]
- He L, Huang X, Kanisicak O, et al. Preexisting endothelial cells mediate cardiac neovascularization after injury. J Clin Invest 2017;127:2968-81. [Crossref] [PubMed]
- Rosenblatt-Velin N, Badoux S, Liaudet L. Pharmacological Therapy in the Heart as an Alternative to Cellular Therapy: A Place for the Brain Natriuretic Peptide? Stem Cells Int 2016;2016:5961342.
- Katz MG, Fargnoli AS, Kendle AP, et al. The role of microRNAs in cardiac development and regenerative capacity. Am J Physiol Heart Circ Physiol 2016;310:H528-41. [Crossref] [PubMed]
- Iaconetti C, Sorrentino S, De Rosa S, et al. Exosomal miRNAs in Heart Disease. Physiology (Bethesda) 2016;31:16-24. [Crossref] [PubMed]
- Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007;129:303-17. [Crossref] [PubMed]
- Rao PK, Toyama Y, Chiang HR, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 2009;105:585-94. [Crossref] [PubMed]
- Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006;38:228-33. [Crossref] [PubMed]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214-20. [Crossref] [PubMed]
- Liu N, Williams AH, Kim Y, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A 2007;104:20844-9. [Crossref] [PubMed]
- Liu N, Bezprozvannaya S, Williams AH, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 2008;22:3242-54. [Crossref] [PubMed]
- Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009;119:2772-86. [Crossref] [PubMed]
- Morton SU, Scherz PJ, Cordes KR, et al. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A 2008;105:17830-5. [Crossref] [PubMed]
- Deacon DC, Nevis KR, Cashman TJ, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development 2010;137:1887-96. [Crossref] [PubMed]
- Miyasaka KY, Kida YS, Banjo T, et al. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech Dev 2011;128:18-28. [Crossref] [PubMed]
- Fish JE, Wythe JD, Xiao T, et al. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011;138:1409-19. [Crossref] [PubMed]
- Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol 2000;223:169-80. [Crossref] [PubMed]
- Chiavacci E, Dolfi L, Verduci L, et al. MicroRNA 218 mediates the effects of Tbx5a over-expression on zebrafish heart development. PLoS One 2012;7:e50536 [Crossref] [PubMed]
- Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261-71. [Crossref] [PubMed]
- Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008;15:272-84. [Crossref] [PubMed]
- Aguirre A, Montserrat N, Zacchigna S, et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 2014;15:589-604. [Crossref] [PubMed]
- Yin VP, Lepilina A, Smith A, et al. Regulation of zebrafish heart regeneration by miR-133. Dev Biol 2012;365:319-27. [Crossref] [PubMed]
- Mohamed TMA, Ang YS, Radzinsky E, et al. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018;173:104-16.e12. [Crossref] [PubMed]
- le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007;26:3699-708. [Crossref] [PubMed]
- Liu X, Cheng Y, Zhang S, et al. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res 2009;104:476-87. [Crossref] [PubMed]
- Liu X, Xiao J, Zhu H, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab 2015;21:584-95. [Crossref] [PubMed]
- Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008;132:875-86. [Crossref] [PubMed]
- Chen J, Huang ZP, Seok HY, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 2013;112:1557-66. [Crossref] [PubMed]
- Shi J, Bei Y, Kong X, et al. miR-17-3p Contributes to Exercise-Induced Cardiac Growth and Protects against Myocardial Ischemia-Reperfusion Injury. Theranostics 2017;7:664-76. [Crossref] [PubMed]
- Tian Y, Liu Y, Wang T, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med 2015;7:279ra38 [Crossref] [PubMed]
- Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376-81. [Crossref] [PubMed]
- Shin CH, Liu ZP, Passier R, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 2002;110:725-35. [Crossref] [PubMed]
- Xiao J, Liu H, Cretoiu D, et al. miR-31a-5p promotes postnatal cardiomyocyte proliferation by targeting RhoBTB1. Exp Mol Med 2017;49:e386 [Crossref] [PubMed]
- Kuehbacher A, Urbich C, Zeiher AM, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 2007;101:59-68. [Crossref] [PubMed]
- Suarez Y, Fernandez-Hernando C, Yu J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A 2008;105:14082-7. [Crossref] [PubMed]
- Suarez Y, Fernandez-Hernando C, Pober JS, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 2007;100:1164-73. [Crossref] [PubMed]
- Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009;324:1710-3. [Crossref] [PubMed]
- Iaconetti C, Polimeni A, Sorrentino S, et al. Inhibition of miR-92a increases endothelial proliferation and migration in vitro as well as reduces neointimal proliferation in vivo after vascular injury. Basic Res Cardiol 2012;107:296. [Crossref] [PubMed]
- Hinkel R, Penzkofer D, Zuhlke S, et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 2013;128:1066-75. [Crossref] [PubMed]
- Wu W, Xiao H, Laguna-Fernandez A, et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation 2011;124:633-41. [Crossref] [PubMed]
- Bang C, Fiedler J, Thum T. Cardiovascular importance of the microRNA-23/27/24 family. Microcirculation 2012;19:208-14. [Crossref] [PubMed]
- Fiedler J, Jazbutyte V, Kirchmaier BC, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011;124:720-30. [Crossref] [PubMed]
- Meloni M, Marchetti M, Garner K, et al. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol Ther 2013;21:1390-402. [Crossref] [PubMed]
- Icli B, Wara AK, Moslehi J, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res 2013;113:1231-41. [Crossref] [PubMed]
- Yin KJ, Olsen K, Hamblin M, et al. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J Biol Chem 2012;287:27055-64. [Crossref] [PubMed]
- Spinetti G, Fortunato O, Caporali A, et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res 2013;112:335-46. [Crossref] [PubMed]
- Fasanaro P, D'Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 2008;283:15878-83. [Crossref] [PubMed]
- Hu S, Huang M, Li Z, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation 2010;122:S124-31. [Crossref] [PubMed]
- Rochette L, Zeller M, Cottin Y, et al. Growth and differentiation factor 11 (GDF11): Functions in the regulation of erythropoiesis and cardiac regeneration. Pharmacol Ther 2015;156:26-33. [Crossref] [PubMed]
Cite this article as: Li N, Rochette L, Rosenblatt-Velin N. Heart regeneration after myocardial infarction: the role of microRNAs. Non-coding RNA Investig 2018;2:26.


