
How T cells go rogue in the absence of Roquins
Roquin-1 and Roquin-2 are RNA-binding proteins (RBPs) essential for modulating T cell activity. Indeed, Roquin dysfunction has been linked to autoimmunity in mice. Essig and colleagues (2017) determine their functions in Foxp3+ T regulatory cells and uncover novel mechanisms of Roquin-mediated regulation of its target mRNAs (1).
Roquin-1 and 2 (encoded by Rc3h1 and Rc3h2, respectively) are RBPs that redundantly regulate mRNA stability and localize to cytoplasmic processing bodies (P-bodies) and stress granules (2-4). Rc3h1 was originally discovered as a suppressor of autoimmunity through an ethylnitrosourea-based forward genetic screen in mice. An informative mouse strain called sanroque harbored a missense Methionine199 to Arginine mutation (M199R) in the Roquin domain of Roquin-1 and resulted in a severe lupus-like syndrome (5). Since Roquin-2 was intact in the sanroque mouse, this mutation likely functions through an autosomal dominant negative mechanism that is not yet completely understood.
Early work demonstrated that Roquins inhibit inducible co-stimulatory (Icos) receptor signaling in T cells through two independent pathways. First, Roquins directly bind the 3' untranslated region (UTR) of Icos mRNA and promote its decay through the Ccr4-Caf1-Not deadenylation complex and the Rck/Edc4/Dcp1a decapping complex (2,3,5-7). In addition, Roquins can bind Ago2 and downregulate the levels of miR-146a while simultaneously targeting Icos mRNA through a target-mediated microRNA decay (TMMD) (8). Augmented Icos expression appears sufficient to stimulate phosphoinositide 3-kinase (PI3K) signaling, phosphatidylinositol 3,4,5-tris-phosphate (PIP3) production and Akt-mediated inactivation of Forkhead Box O1 (Foxo1), a transcription factor required for T cell suppression, by promoting its export from the nucleus to the cytoplasm (9,10). Roquins regulate additional mRNA targets including Tnfa in macrophages and Ox40 in T cells (2,11) through binding to structured stem loop (SL) elements such as the trinucleotide loop-containing constitutive decay element (CDE) first described in the 3'UTR of Tnfa mRNA (11) and the hexanucleotide loop-containing alternative decay element (ADE) identified from a systematic evolution of ligands by exponential enrichment (SELEX) screen and shown to be present in the 3'UTR of Ox40 mRNA (12). Roquins were also shown to regulate the levels of several mRNAs that encode components of the NF-κB pathway including A20, NFKBID and NFKBIZ using Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP) (13) in human embryonic kidney 293 (HEK293) cells (7).
Roquins contain a really interesting new gene (RING) domain followed by a Roquin domain, a C3H1-type zinc finger domain and a proline rich domain at the C-terminus. The Roquin domain is a winged-helix fold that mediates Roquin’s binding to its target mRNAs (14-16) together with flanking higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains (8). The C-terminus recruits the Ccr4-Caf1-Not complex to the mRNA (11). Whether the zinc finger domain contributes to Roquin’s RNA binding activity is currently unknown. The RING domain is an E3 ubiquitin ligase that autoubiquitinates Roquins (17). While an in vivo requirement for Roquin-2’s RING domain was not immediately apparent, germline deletion of Roquin-1’s RING domain resulted in perinatal lethality (18). The RING domain was proposed to downregulate 5' adenosine monophosphate-activated protein kinase α1 (Ampkα1) levels through a protein-protein interaction independently of its mRNA binding activity (17) suggesting multiple modes of Roquin function. In turn, hypoactive Ampkα1 causes increased mTOR signaling and mRNA translation leading to enhanced T follicular helper (Tfh) cell differentiation. However, in mouse embryonic fibroblasts (MEFs), Essig et al. found that the RING domain was not required for Roquin-mediated inhibition of protein biosynthesis (1). Further work is needed to address this discrepancy.
Whether Roquins regulate additional cellular pathways to mediate their immunosuppressive functions has been an outstanding question. In a recent study, Essig et al. (1) identify novel targets through which Roquins orchestrate T-helper cell differentiation (Figure 1). By employing PAR-CLIP (13) in MEFs, the authors discovered two additional mRNA targets, itchy E3 ubiquitin protein ligase (Itch) and Phosphatase and tensin homolog (Pten). Fortunately, these two transcripts happen to be expressed in T cells as well, and play prominent regulatory roles in T cells. Intriguingly, in the absence of Roquins these two mRNA targets display opposing fates. While Roquins impair the expression of Icos and Itch, consistent with the idea that the Ccr4-Caf1-Not complex is recruited to the mRNAs; Roquins appear to promote the levels of Pten mRNA (Figure 1). Itch is an E3 ubiquitin ligase that has been shown to ubiquitinate and promote the degradation of Foxo1 by the proteasome (19). By inhibiting Itch expression, Roquins mediate their T cell suppressive activity through the maintenance of Foxo1 levels (1). However, this is not necessarily sufficient as Foxo1 can be inhibited via Akt phosphorylation which promotes its nuclear export to the cytoplasm.
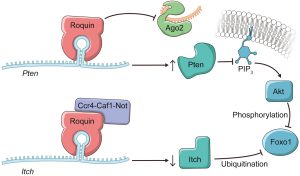
To circumvent this pathway, Roquins also increased Pten mRNA and Pten protein levels modestly. Pten indirectly represses Akt activity by dephosphorylating PIP3. As a result, Pten indirectly promotes the nuclear localization of Foxo1 and its T cell suppressive gene expression program. Since the Roquin binding site overlaps the miR-17-5p binding site in Pten mRNA 3'UTR (Figure 2), Essig and colleagues proposed a novel mechanism of Roquin activity by competing with miR-17-5p for binding to Pten mRNA. The absence of Roquin expression in T cells enhanced the association of Pten mRNA with Ago2 and subsequent miRNA-mediated repression (1). It remains to be determined whether Roquins regulate additional mRNA targets through this novel mechanism.
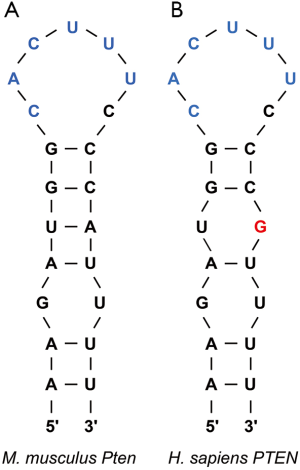
In sum, Roquins function as a key regulatory hub of T cell activity by orchestrating multiple independent pathways that control Foxo1 levels and subcellular localization. It will be important to systematically identify Roquin’s targets transcriptome-wide and determine the fates of those mRNAs. If additional examples are validated, then how Roquins mediate opposing effects on their mRNA targets will be an interesting question. It seems that Roquins do not always recruit the Ccr4-Caf1-Not complex to their target mRNAs. It is also plausible that Roquins interact with additional cofactors that contribute to determining the mRNA target’s fate upon Roquin binding. Indeed, the same group has recently reported that Roquin-1 interacts with nuclear fragile X mental retardation protein interacting protein 2 (Nufip2) which enhances the binding affinity of Roquin-1 to the 3'UTR of Icos mRNA (20). An alternative possibility might be subtle differences in the stem-loop structure. While the ADE and CDE are hexanucleotide and trinucleotide loops respectively, the proposed Roquin-binding Pten loop is heptanucleotide. Furthermore, it remains to be identified whether Roquins regulate T cell activity via pathways other than PI3K-mTOR signaling. It would be worthwhile repeating the PAR-CLIP experiment in T cells as it can serve as a powerful approach to explore the binding of Roquins to additional mRNA targets that encode factors involved in T cell activation.
Given their conservation and ubiquitous expression, another outstanding question is whether Roquins regulate gene expression programs post-transcriptionally in other tissues or cell types. Roquin interacts with the Ccr4-Not complex in D. melanogaster, an organism that lacks T cells (21). In addition, Roquins are expressed in mouse non-immune tissues such as the lung and heart (2). Germline deletion of Rc3h1 results in curly tails, defects in the caudal spinal column, malformed alveoli in the lung, and perinatal lethality (≤6 h after birth) (22). This raises the possibility of a Roquin function outside the immune system. For instance, Roquin upregulation has been recently correlated with poor survival in breast cancer patients (23).
Since the Methionine199 residue is conserved in humans, the body of work in mice predicts that human patients suffering from familial autoimmune disorders may harbor Roquin mutations perhaps similar if not identical to the sanroque mutation. It is also conceivable that autoimmunity could result from CDE-disrupting mutations within the 3'UTR of Icos, Itch or other Roquin targets. It is worth noting that blockade of Interferon-γ was shown to ameliorate the lupus-like disease in sanroque mice and thus is a promising therapeutic avenue for future exploration (24). Ultimately, a better understanding of the molecular mechanisms of Roquin function will allow the field to identify additional therapeutic targets to alleviate Roquin-mediated disorders. The clues learnt from studying Roquin supports the general notion that viewing the immune system through the lens of RBPs will help shed new light on important regulatory circuits (25).
Acknowledgments
We thank Bryan Chim (NIAID) for discussions on PAR-CLIP data analysis, Ryan Kissinger (NIAID) for graphic art in Figure 1, and Saifeng Wang for critically reading this editorial.
Funding: Our work was supported by the Intramural Research program of the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Yuan Xie (Shanghai Tongji Hospital, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.03.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Essig K, Hu D, Guimaraes JC, et al. Roquin Suppresses the PI3K-mTOR Signaling Pathway to Inhibit T Helper Cell Differentiation and Conversion of Treg to Tfr Cells. Immunity 2017;47:1067-82.e12. [Crossref] [PubMed]
- Vogel KU, Edelmann SL, Jeltsch KM, et al. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity 2013;38:655-68. [Crossref] [PubMed]
- Glasmacher E, Hoefig KP, Vogel KU, et al. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol 2010;11:725-33. [Crossref] [PubMed]
- Athanasopoulos V, Barker A, Yu D, et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J 2010;277:2109-27. [Crossref] [PubMed]
- Vinuesa CG, Cook MC, Angelucci C, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 2005;435:452-8. [Crossref] [PubMed]
- Yu D, Tan AH, Hu X, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 2007;450:299-303. [Crossref] [PubMed]
- Murakawa Y, Hinz M, Mothes J, et al. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat Commun 2015;6:7367. [Crossref] [PubMed]
- Srivastava M, Duan G, Kershaw NJ, et al. Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis. Nat Commun 2015;6:6253. [Crossref] [PubMed]
- Stone EL, Pepper M, Katayama CD, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity 2015;42:239-51. [Crossref] [PubMed]
- Kerdiles YM, Stone EL, Beisner DR, et al. Foxo transcription factors control regulatory T cell development and function. Immunity 2010;33:890-904. [Crossref] [PubMed]
- Leppek K, Schott J, Reitter S, et al. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 2013;153:869-81. [Crossref] [PubMed]
- Janowski R, Heinz GA, Schlundt A, et al. Roquin recognizes a non-canonical hexaloop structure in the 3′-UTR of Ox40. Nat Commun 2016;7:11032. [Crossref] [PubMed]
- Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129-41. [Crossref] [PubMed]
- Schlundt A, Heinz GA, Janowski R, et al. Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nat Struct Mol Biol 2014;21:671-8. [Crossref] [PubMed]
- Schuetz A, Murakawa Y, Rosenbaum E, et al. Roquin binding to target mRNAs involves a winged helix-turn-helix motif. Nat Commun 2014;5:5701. [Crossref] [PubMed]
- Tan D, Zhou M, Kiledjian M, et al. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat Struct Mol Biol 2014;21:679-85. [Crossref] [PubMed]
- Ramiscal RR, Parish IA, Lee-Young RS, et al. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. Elife 2015;4: [Crossref] [PubMed]
- Pratama A, Ramiscal RR, Silva DG, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 2013;38:669-80. [Crossref] [PubMed]
- Xiao N, Eto D, Elly C, et al. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol 2014;15:657-66. [Crossref] [PubMed]
- Rehage N, Davydova E, Conrad C, et al. Binding of NUFIP2 to Roquin promotes recognition and regulation of ICOS mRNA. Nat Commun 2018;9:299. [Crossref] [PubMed]
- Sgromo A, Raisch T, Bawankar P, et al. A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat Commun 2017;8:14307. [Crossref] [PubMed]
- Bertossi A, Aichinger M, Sansonetti P, et al. Loss of Roquin induces early death and immune deregulation but not autoimmunity. J Exp Med 2011;208:1749-56. [Crossref] [PubMed]
- Ramanathan M, Majzoub K, Rao DS, et al. RNA-protein interaction detection in living cells. Nat Methods 2018;15:207-12. [Crossref] [PubMed]
- Lee SK, Silva DG, Martin JL, et al. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity 2012;37:880-92. [Crossref] [PubMed]
- Yuan J, Muljo SA. Exploring the RNA world in hematopoietic cells through the lens of RNA-binding proteins. Immunol Rev 2013;253:290-303. [Crossref] [PubMed]
Cite this article as: Akef A, Muljo SA. How T cells go rogue in the absence of Roquins. Non-coding RNA Investig 2018;2:20.


