
Long non-coding RNA in the control of genome stability and cancer phenotypes
Our genomes are continuously under remodeling. Variations at the single-nucleotide level, named single-nucleotide polymorphisms (SNPs) when present in more than 1% of the population, drive evolution and are crucial for diversity within the population and different susceptibility to diseases. SNPs within an intergenic region at 11q13 are associated with breast cancer risk (1,2). Although until recently it was not widely accepted that variations within regions with no protein-coding potential could be associated with diseases, we are now aware that the vast majority of the genome is transcribed into non-coding RNA (ncRNA) molecules (3), which are involved in a variety of biological processes. Supporting the functional role of the non-coding transcriptome, mutations within the non-coding genome are associated with cancer and other human diseases (4). Among the ncRNAs, long ncRNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides with no protein-coding potential, although recently small peptides encoded by lncRNAs have been identified (5,6). LncRNAs mainly localize in the nucleus, where they regulate gene expression and other cellular processes by interacting with the cellular macromolecules DNA, RNA, and proteins (7,8). The lncRNAs-mediated control of gene expression is achieved both at the transcriptional and at the post-transcriptional level by modulation of the chromatin status and/or by directly interacting with functional proteins or RNAs (7,8). Through each one of these mechanisms lncRNAs can control several cellular functions, including cell cycle, cell proliferation, apoptosis, and DNA damage response (DDR), all pathways which dysregulation has been associated with cancer (9). As a consequence, aberrant lncRNAs expression can drive cancer phenotypes. For example, the lncRNA HOTAIR (HOX transcript antisense RNA), which controls chromatin status by binding and targeting to chromatin the Polycomb repressive complex PRC2 (10), promotes cancer invasiveness and metastasis by reprogramming the chromatin status and, thus, altering gene expression (11). Similarly, SChLAP1 (second chromosome locus associated with prostate-1) antagonizes the genome-wide localization and regulatory functions of the SWI/SNF (SWItch/Sucrose non-fermentable) chromatin-modifying complex and contributes to prostate cancer progression (12). Another way lncRNAs control cellular processes is by directly targeting RNAs and modulating their splicing, stability, and translation. LncRNA-mediated induction of alternative splicing and following inclusion of an internal ribosome entry site is the mechanism leading to the expression of an E-cadherin repressor during epithelial-mesenchymal transition (13). In addition to binding DNA and RNA, lncRNAs can also bind proteins and modulate their function. This is the case, for example, for the lncRNAs SAMMSON (survival associated mitochondrial melanoma-specific oncogenic noncoding RNA), which affects mitochondrial function in a pro-oncogenic way in melanoma by interacting with the mitochondrial regulator protein p32 (14). This is also one of the ways lncRNAs control genome stability, one of the hallmarks of cancer (9). Every day thousands of lesions challenge the stability of our genomes (15). DNA double-strand breaks (DSBs) are among the most dangerous DNA lesions that, if not properly repaired, can lead to genomic instability, which is associated with cancer initiation/progression and ageing. In order to cope with DNA lesions, cells have evolved a signaling cascade named DDR, which promptly recognizes the lesions, signals their presence, and in turn promotes efficient repair or, alternatively, cell death and cellular senescence (16). DSBs are mainly repaired by simply re-joining of the two DNA ends, a process known as non-homologous end joining (NHEJ), or by homologous recombination (HR), in which homologous sequences are used as template to copy back the information missing at the damaged locus (17). Recently, RNA has been discovered as a new player in the DNA damage signaling and repair world. Small ncRNA molecules, named DDRNAs DNA (damage response RNAs) and diRNAs (damage-induced RNAs), contribute, respectively, to DNA damage signaling and repair by promoting the recruitment of DNA damage proteins to the site of damage (18,19). More recently, the generation of these small RNA molecules has been integrated in a larger picture. It has been demonstrated that RNA polymerase II can bind DNA ends of DSBs where it transcribes lncRNA molecules, named damage-induced lncRNAs (dilncRNAs). DilncRNAs are processed to generate DDRNAs and contribute to DNA damage signaling and repair by forming a scaffold for their recruitment to the damaged DNA (20). In line with these emerging findings, nascent RNAs at DSBs located in actively transcribed genes interact with DNA damage proteins and contribute to their recruitment to the site of damage (21). Similarly, the lncRNA LINP1 (lncRNA in non-homologous end joining pathway 1) controls NHEJ mediated-DSB repair by providing a scaffold linking NHEJ proteins (22). Another example is the lncRNA DDSR1 (DNA damage-sensitive RNA1), which modulates HR-mediated repair by binding hnRPUL1, a factor already known to control DNA end resection (23), and in this way modulating BRCA1 recruitment to DSBs (24). However, DDSR1 not only contributes to genome stability by binding DNA damage proteins, but it also controls the expression of DNA damage related-genes. This is one of the major mechanisms through which lncRNAs modulate genome stability. For example, the lncRNA DINO (damage induced non-coding) contributes to the amplification of the cellular response to DNA damage by binding and stabilizing p53 and, therefore, controlling the expression of its target genes (25). Differently, lincRNAp21 modulates DNA damage response by interacting with and controlling the localization of the ribonucleoprotein hnRNP-K to promoters of p53 target genes (26). Another example is the lncRNA NORAD (ncRNA activated by DNA damage), which controls the expression of DNA repair genes by sequestering negative regulators of their expression (27). By performing RNA CaptureSeq on breast cancer cell lines, Betts and colleagues identify two lncRNAs, CUPID1 and CUPID2 (CCND1-uprstream intergenic DNA repair 1 and 2), which share a bi-directional promoter and are transcribed from the breast cancer-risk locus 11q13. By chromatin conformation capture, they demonstrate that the enhancer PRE1 interacts with the predicted CUPID1 and CUPID2 bi-directional promoter and controls their expression. Importantly, two of the breast cancer associated SNPs within the cancer-risk locus 11q13 are located within the enhancer and reduce its activity on CUPID1 promoter by disrupting the enhancer/promoter interaction (Figure 1), as demonstrated by chromatin conformation capture in cells heterozygous for the cancer-risk SNP. Gene expression studies reveal that CUPID1 and CUPID2 control the expression of genes involved in several physiological processes, including DNA replication, recombination, and repair. Indeed, CUPID1 and CUPID2 depletion directs repair toward NHEJ, an error-prone pathway, while reducing HR-mediated repair (Figure 1). Accordingly, breast tumors with low CUPID1 and CUPID2 expression have an HR mutation signature and high levels of structural variants. Intriguingly, while CUPID2 is localized in the cytoplasm and in the nucleus, CUPID1 has a nuclear localization only. It cannot be excluded, therefore, that these newly identified lncRNAs not only contribute to genome stability by modulating the expression of DNA damage response proteins but could also actually localize to DNA damage sites and directly contribute to DNA damage signaling and repair.
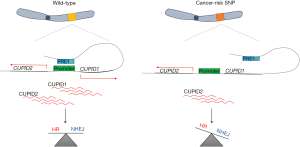
This finding is crucial since lncRNAs are being currently used as predictors or prognostic markers for cancer and could also be valid targets for cancer therapy (25). Tremendous improvements in the RNA-targeting therapeutics field have been recently obtained. For examples, antisense oligonucleotides (ASOs), locked nucleic acids which avidly bind their target RNA and block their function, have been successfully used for targeting mRNAs and ncRNAs involved in a variety of diseases (28). Moreover, in the future, the recently discovered CRISPR-mediated RNA targeting could also be exploited in this field (29,30). In conclusion, the identification of the lncRNAs CUPID1 and CUPD2 not only helps understanding how SNPs within the 11q13 locus are associated with cancer, but also offers additional opportunities for cancer therapy.
Acknowledgments
Funding: Work in Fabrizio d’Adda di Fagagna’s laboratory is supported by the Associazione Italiana per la Ricerca sul Cancro, AIRC (application 12971), Human Frontier Science Program (contract RGP 0014/2012), Cariplo Foundation (grant 2010.0818 and 2014-0812), Marie Curie Initial Training Networks [FP7 PEOPLE 2012 ITN (CodAge)], Fondazione Telethon (GGP12059), Association for International Cancer Research (AICR-Worldwide Cancer Research Rif. N. 14-1331), Progetti di Ricerca di Interesse Nazionale (PRIN) 2010–2011, the Italian Ministry of Education Universities and Research EPIGEN Project, a European Research Council advanced grant (322726) and AriSLA (project ‘DDRNA and ALS’).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Jinzhe Zhou (Department of General Surgery, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.03.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 2010;42:504-7. [Crossref] [PubMed]
- French JD, Ghoussaini M, Edwards SL, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet 2013;92:489-503. [Crossref] [PubMed]
- Clark MB, Amaral PP, Schlesinger FJ, et al. The reality of pervasive transcription. PLoS Biol 2011;9:e1000625; discussion e1102.
- Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012;337:1190-5. [Crossref] [PubMed]
- Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160:595-606. [Crossref] [PubMed]
- Nelson BR, Makarewich CA, Anderson DM, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016;351:271-5. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Prensner JR, Iyer MK, Sahu A, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013;45:1392-8. [Crossref] [PubMed]
- Beltran M, Puig I, Pena C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 2008;22:756-69. [Crossref] [PubMed]
- Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016;531:518-22. [Crossref] [PubMed]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000;65:127-33. [Crossref] [PubMed]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179-204. [Crossref] [PubMed]
- Ceccaldi R, Rondinelli B, D'Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol 2016;26:52-64. [Crossref] [PubMed]
- d'Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol 2014;24:171-8. [Crossref] [PubMed]
- D'Alessandro G, d'Adda di Fagagna F. Transcription and DNA Damage: Holding Hands or Crossing Swords? J Mol Biol 2017;429:3215-29. [Crossref] [PubMed]
- Michelini F, Pitchiaya S, Vitelli V, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol 2017;19:1400-11. [Crossref] [PubMed]
- Chakraborty A, Tapryal N, Venkova T, et al. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat Commun 2016;7:13049. [Crossref] [PubMed]
- Zhang Y, He Q, Hu Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol 2016;23:522-30. [Crossref] [PubMed]
- Polo SE, Blackford AN, Chapman JR, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Molecular cell 2012;45:505-16. [Crossref] [PubMed]
- Sharma V, Khurana S, Kubben N, et al. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep 2015;16:1520-34. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell 2016;29:452-63. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Lee S, Kopp F, Chang TC, Sataluri A, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016;164:69-80. [Crossref] [PubMed]
- McClorey G, Wood MJ. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol 2015;24:52-8. [Crossref] [PubMed]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature 2017;550:280-4. [Crossref] [PubMed]
- Strutt SC, Torrez RM, Kaya E, et al. RNA-dependent RNA targeting by CRISPR-Cas9. Elife 2018;7:e32724 [Crossref] [PubMed]
Cite this article as: D’Alessandro G, d’Adda di Fagagna F. Long non-coding RNA in the control of genome stability and cancer phenotypes. Non-coding RNA Investig 2018;2:13.


