
A link between a synonymous SNP and the clinical response to tyrosine kinase inhibitors
Epidermal growth factor receptor (EGFR) is a member of the large receptor tyrosine kinase (RTK) family. Upon binding to corresponding growth factor ligands, EGFR dimerizes, and triggers intrinsic protein tyrosine kinase activity that regulates cell growth, proliferation, differentiation and survival (1,2). EGFR consists of an extracellular ligand-binding domain, a single transmembrane region and an intracellular tyrosine kinase domain (3). Phosphorylated EGFR stimulates many downstream signaling pathways by recruitment of effector proteins. The three major pathways are (I) Ras/Raf /MAPK pathway, (II) PI3K/Akt pathway and (III) JAK/STAT pathway, all of which are well known for their roles in the regulation of fundamental cellular processes (4).
While EGFR-mediated signaling is required for normal cellular function, the activity of EGFR is frequently dysregulated in cancer through various mechanisms, such as overexpression, mutation and deletion. For instance, more than 50% of the studied cancer types, including bladder, ovarian, breast and colorectal cancer, are associated with increased expression of EGFR (5); early retrospective analyses showed EGFR overexpression in 62% of non-small cell lung cancer (NSCLC) cases, which is associated with poor prognosis (6). Overexpression of EGFR increases cell exposure to the related ligands, such as epidermal growth factor (EGF) and transforming growth factor-α (TGF-α), and subsequently upregulates cellular activities. Activating mutations in the EGFR gene, such as deletions in exon 19 and mutation L858R in exon 21, can destabilize the auto-inhibited conformation of EGFR through steric alterations, leading to constitutively active signaling cascades even in the absence of the ligands (7). Tumors with increased EGFR activity can often benefit from treatment with EGFR-tyrosine kinase inhibitors (TKIs).
Indeed, EGFR-TKIs have been used as a standard treatment in the clinic for patients carrying mutant EGFR in many types of cancer. The first generation TKIs such as gefitinib (8) and erlotinib (9) can bind the kinase domain and compete with ATPs, thus inhibiting phosphorylation of the kinase domain. However, approximately 50% of the patients may develop resistance to the first generation TKIs after treatment, due to selection of clones with secondary mutation T790M (10). It was proposed that the T790M mutation on the kinase domain increases the affinity to ATPs, making ATPs one order of magnitude more competitive to the reversible TKIs (11). Thus, new generations of TKIs have been developed to tackle this type of resistance. Nevertheless, the first generation of TKIs still plays a major role in treatment of cancer with active EGFR.
Hence, predictive biomarkers such as activating mutations, especially in exons 18–21 of EGFR or EGFR amplification, are critical to identify patients suitable for TKIs, and testing for these mutations and tailoring therapy have been accepted as standard clinical practice for various types of cancer. However, although there is evidence that head and neck squamous-cell carcinoma (HNSCC) can be dependent on EGFR signaling, these markers do not seem to show any benefit for oral squamous-cell carcinoma (OSCC), a subgroup of HNSCC. However, Tan et al. identified two of OSCC patients who showed dramatic responses to gefitinib (12). Further characterization revealed a synonymous homozygous single-nucleotide variant (or single nucleotide polymorphism, SNP) at position 2361 of the coding region (rs10251977; 2361G>A) in EGFR exon 20 (13). In A/A or G/A genotype, the level of a long non-coding RNA (lncRNA), called EGFR-AS1, is decreased, whereas EGFR isoform D/A ratio is increased as compared to G/G genotype (Figure 1). A consequence of these changes is increased sensitivity to TKIs such as gefitinib.
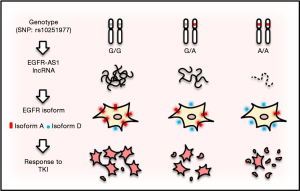
In this recent paper (13), the authors used primary cell culture derived from the patients, and demonstrated that the heterozygous SNP (G/A) is more resistant to TKIs than the homozygous A/A counterpart, and the homozygous wild type (G/G) cells such as NCC-HN1 showed the highest resistance among them. Moreover, the cells with G/G genotype revealed a higher level of phosphorylated EGFR, AKT, ERK and ribosomal protein S6 (S6) than the cells with A/A genotype in response to gefitinib and erlotinib, respectively, suggesting that SNP G>A may impact the TKI response through these pathways. The function of this alteration (G>A) was elegantly demonstrated by knockin of G/A in the G/G background of NCC-HN1 cells. For instance, the edited G/AAAV cells were more sensitive to gefitinib with a lower level of phosphorylation of these target proteins than the G/GAAV cells. Experiments with siRNA or LNA antisense oligonucleotides further support the role of G/A in response to TKIs in OSCC. These results provide evidence that a synonymous SNP is functional, and can serve as a driving force for increased sensitivity to TKIs and phosphorylation of EGFR, AKT, ERK and S6. Of interest, this sensitivity is specific to inhibitors targeting the kinase domain such as gefitinib, erlotinib, afatinib, dacomitinib, lapatinib, but has not been seen with monoclonal antibodies against the extracellular domain of EGFR, such as cetuximab, suggesting that a unique mechanism is involved.
Now a question is how a silent (synonymous) SNP that does not change amino acid sequence (EGFR-Q787Q) would cause a better clinical outcome. Since the alteration also involves lncRNA EGFR-AS1, which is transcribed from the same region but in an opposite direction (Figure 2A), it would not be surprised that this SNP may impact expression of EGFR-AS1. Indeed, quantitative RT-PCR assays indicated that a lower EGFR-AS1 level is associated with this SNP and these cells are more sensitive to TKIs. This downregulation of EGFR-AS1 in the A/A genotype cells is due to its decreased stability. Moreover, the function of EGFR-AS1 regulating TKI resistance was further validated by knockdown using small interfering RNA (siRNA), sensitizing the previously resistant cell line (G/G) to TKIs.
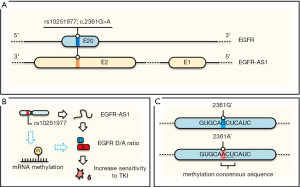
Mechanistically, the same approaches (knockdown of EGFR-AS1 by siRNA or LNA oligo, AAV-mediated generation of G/A genotype) revealed the upregulation of EGFR isoform D/A ratio. For instance, this ratio was particularly higher in the sensitive cell lines (A/A or G/A) than in the resistant line (G/G). EGFR isoform D has been previously shown to be associated with increased sensitivity to TKIs (14) although the underlying mechanism is not fully understood. The effect of EGFR isoform D on TKI sensitivity was demonstrated by knockdown of isoform D, but not isoform A, resulting in gain of resistance to TKI in previously sensitive line (A/A). Finally, this correlation between this SNP (G/G vs. A/A) and clinical response was further confirmed in a cohort of additional 8 patients (6 AAs vs. 2 GGs), highlighting the significance of these findings.
It appears clear that G2361A-mediated increased level of EGFR isoform D is responsible for the increased sensitivity to TKIs, and EGFR-AS1 may contribute to the formation of EGFR isoform D as the authors proposed. The role of lncRNAs in gene splicing has been well documented in the literature and a well-known example is MALAT1 that can interact with splicing factors and influence their distribution in nuclear speckle domains, leading to different splicing patterns (15). Furthermore, lncRNAs can also regulate gene splicing through their encoded short peptides. For example, a short peptide derived from lncRNA HOXB-AS3 can interact with heterogeneous ribonucleoprotein A1 (hnRNP A1) and promote the expression of isoform I of pyruvate kinase (PKM1) (16). If EGFR-AS1 is involved in regulation of EGFR isoform D, it is likely that EGFR-AS1 serves a negative regulator because in the A/A genotype cells, EGFR isoform D is increased with a decreased level of EGFR-AS1.
In this regard, we would like to propose an additional possibility that A/A genotype may lead to upregulation of EGFR isoform D (Figure 2B,C). Although it is well known that DNA methylation plays a critical role in gene expression, accumulating evidence indicates that RNA can also be reversibly methylated. Many studies support the role of RNA methylation in the post-transcriptional regulation of gene expression. RNA methylation on the A base (m6A) is the most abundant posttranscriptional modification of mammalian mRNA. We note that when G/G is changed to A/A in this case, GCU becomes ACU (Figure 2C) that matches perfectly with the consensus RNA methylation sequence (17). Apparently, RNA methylation (m6A) can change structure of mRNAs that can lead to different RNA-protein interactions. For instance, MALAT1 methylation can enhance its interaction with splicing factors such as heterogeneous nuclear ribonucleoprotein C (hnRNP C). Thus, RNA methylation has been shown to efficiently interact with the target RNAs, and affect alternative splicing and abundance of multiple target mRNAs (18). Of course, other possible functions of m6A of EGFR regarding rs10251977 cannot be ruled out even if it is indeed proven that this SNP enhances EGFR methylation.
In summary, the study by Tan et al. provides an excellent example that a synonymous SNP can alter EGFR addiction, where lncRNA EGFR-AS1 and EGFR isoform D are important players in this pathway. Future work should focus on how to incorporate these findings into the biomarker discovery pathway and to better understand how SNP, lncRNA and EGFR isoforms interact to impact clinical outcomes.
Acknowledgments
Funding: This work was in part supported by NIH grant R01 CA154989 (YM).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Yikun Yao (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.01.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006;7:505-16. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer 2013;13:663-73. [Crossref] [PubMed]
- Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B 2015;5:390-401. [Crossref] [PubMed]
- Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer 2001;37:S9-15. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Dixit A, Verkhivker GM. Structure-functional prediction and analysis of cancer mutation effects in protein kinases. Comput Math Methods Med 2014;2014:653487
- Mitsudomi T, Morita S, Yatabe Y, et al. Updated overall survival results of WJTOG 3405, a randomized phase III trial comparing gefitinib (G) with cisplatin plus docetaxel (CD) as the first-line treatment for patients with non-small cell lung cancer harboring mutations of the epidermal growth factor receptor (EGFR). J Clin Oncol 2012;30:abstr 7521.
- Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883-9. [Crossref] [PubMed]
- Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol 2012;7:1640-4. [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Tan EH, Goh C, Lim WT, et al. Gefitinib, cisplatin, and concurrent radiotherapy for locally advanced head and neck cancer: EGFR FISH, protein expression, and mutational status are not predictive biomarkers. Ann Oncol 2012;23:1010-6. [Crossref] [PubMed]
- Tan DS, Chong FT, Leong HS, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med 2017;23:1167. [Crossref] [PubMed]
- Albitar L, Pickett G, Morgan M, et al. EGFR isoforms and gene regulation in human endometrial cancer cells. Mol Cancer 2010;9:166. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
- Huang JZ, Chen M, Chen D, et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell 2017;68:171-84.e6. [Crossref] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. [Crossref] [PubMed]
- Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015;518:560-4. [Crossref] [PubMed]
Cite this article as: Peng WX, Fan LT, Mo YY. A link between a synonymous SNP and the clinical response to tyrosine kinase inhibitors. Non-coding RNA Investig 2018;2:6.


