
A new method for the identification of thousands of circular RNAs
Introduction
Circular RNAs (circRNAs) have recently emerged as a novel class of abundant endogenous non-coding RNAs (ncRNAs) with regulatory potential in mammalian cells. Owing to the difficulty to identify these RNAs through traditional methods dedicated to the analysis of linear RNAs, our knowledge of these intriguing RNA species has remained limited. Recent efforts to develop novel biochemical enrichment strategies coupled to deep sequencing allowed their systematic identification for comprehensive studies of their biogenesis and function. The manuscript by Panda et al. discussed herein provides an improvement to existing methods usually based on depletion in polyadenylated RNAs and resistance to linear-RNA exonucleases (1).
Discovery of circRNAs
The existence of circRNAs was first reported in the 1970s in plant viroids and yeast mitochondria (2,3), and later on in higher eukaryotes (4). Very few new circRNAs were identified over the years including circRNAs from the Sry gene in mouse testis (5) or from the cytochrome P450 gene in rats (6) where their levels are correlated with exon skipping. Despite the fact that circRNAs have been detected in human nuclear extracts (7), they have long been considered as byproducts of splicing or debranching of the lariat produced by the debranching enzyme 1 (DBR1), at least over the last two decades (8).
circRNAs: origin, biogenesis and nomenclature
In addition to being considered as “junk” RNA, circRNAs were not identified in traditional sequencing techniques because they cannot be separated on the criterion of size and they do not have, by definition, 5’- and 3’- ends nor poly(A) tails (8). With the advent of high throughput sequencing techniques and the development of dedicated bioinformatic analysis [for review see (9)], many new circRNAs have now been identified and further classified according to their origin and biogenesis. Since the nomenclature of circRNAs can lead to some confusion, we first review the main referenced classes that are depicted on Figure 1. The first class is referred to as circular intronic RNAs (ciRNAs) that, as their name implies, originate from the circularization of intronic sequences. During the splicing reaction, the lariat that is formed may escape debranching by DBR1. The 3’-linear extremity is then trimmed by an exonuclease leading to the formation of a perfect circRNA. A different class is designated as circRNAs that are covalently closed by a mechanism called back-splicing. In canonical linear splicing, the 5’ donor splice site joins the 3’ acceptor splice site to form a 5’-3’ splice junction (consensus sequence AG/GT), combining exons in a sequential order. In contrast, back-splicing goes in the reverse direction. Thus, a donor splice site will react with an acceptor splice site located upstream, allowing the formation of a 3’-5’ backsplice junction with the particular consensus sequence GT/AG. circRNAs that retain introns between two or more exons are called exon-intron circRNAs (EIcircRNAs) as opposed to intronic circRNAs (IcircRNAs) exclusively composed of intronic sequences or exonic circRNAs (EcircRNAs) that contain only exons (1,4).
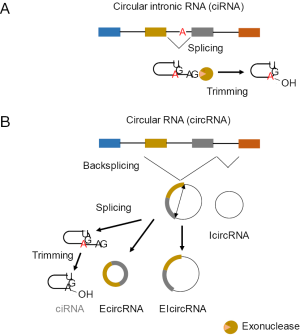
circRNAs are between 100 nt and several kb in size, they originate from both coding and non-coding genes, and their levels are not correlated with expression of the host gene (10). circRNAs are also very stable in the cells with a half-life of more than 48 hours, although not in blood serum (<15 seconds) probably due to the presence of endonucleases. EcircRNAs have been shown to adopt a cytoplasmic localization (4,8) although the export system remains unknown, in contrast to EIcircRNAs that are preferentially located in the nucleus (11). The combination of these features, especially the lack of correlation between the expression of the host genes and the levels of circRNAs, strongly suggests that their release is actively regulated and operates physiological functions in the cells (8).
Biological functions of circRNAs
Many studies have suggested that the exonic sequences carried by circRNAs contain target sequences for microRNAs (miRNAs) and could act as sponges or endogenous competitors for miRNAs (8). This is the case for the circRNA identified in Sry, which contains several binding sites for the mmu-miRNA-138. Overexpression of ectopic circRNA Sry leads to the reduction of the mmu-miRNA-138-mediated knockdown of a luciferase construct containing miRNA-138 binding sites (12). Similarly, the circWDR77, circRNA produced from linear WDR77 transcripts, acts as a sponge for the hsa-miRNA-124 and prevents the knockdown of FGF-2 (10). As previously mentioned, EIcircRNAs represent a particular class of circRNAs localized in the nucleus. They have been shown to regulate transcription in cis through their interaction with the RNA polymerase II, U1 nuclear RNA and the promoter of their host gene. In addition, knockdown of EIcircRNAs can cause a decrease in the mRNA levels of their host genes (11). As it has already been shown that almost all RNAs interact with RNA binding proteins, it is very likely that circRNAs belong to large complexes called circRNPs (13). Indeed, circRNAs can exist in complex with Argonaute (AGO) proteins (8) or with the IGF2BP3 or Insulin-like Growth Factor 2 binding protein 3 (14). It has also been proposed that circRNAs could act as sponges for RBPs or as scaffolds to facilitate the interaction between several RBPs (8).
circRNAs were long considered as ncRNAs because they were not detected in polysomes (5,8,14). However, an unexpected function emerged along which some circRNAs have the ability to code for proteins, or at least peptides. Three publications, summarized by Schneider and Bindereif (15), revealed the translation of circRNAs into small proteins through a cap-independent translation initiation, further confirmed by polysome fractionation and mass spectrometry experiments (16-18). Therefore, circRNAs may possess the ability to act both as coding and ncRNAs, as it has already been reported in the case of the so-called bifunctional RNAs (19).
More generally, several circRNAs have been implicated in development, pluripotency, proliferation, differentiation or migration of normal and tumor cells (16). For example, overexpression of circPTK2 promotes proliferation and migration of bladder cancer cells consistent with its high levels found in these cancer cells (20). circBIRC6 contributes to maintenance of pluripotency in human embryonic stem cells by “sponging” hsa-miRNA-34a and -145, which otherwise target pluripotency genes (21). circWDR77, described above, facilitates proliferation and migration of vascular smooth muscle cells (10).
Given the many physiological functions that circRNAs may operate, and in order to understand their molecular functions, there is a burning need to proceed with their exhaustive identification. This will ultimately allow circRNAs to be used as innovative biomarkers and open up new therapeutic approaches in the treatment of cancer or other human diseases where RNA splicing, and hence the production of circRNAs, is defective.
A new method allows the identification of many new circRNAs
Since most of circRNAs reported to date are the result of backsplice reactions (22) (circBase, http://www.circbase.org), biocomputational strategies developed for their identification were based on the detection of the backsplice signature GT/AG (8). In order to improve their identification experimentally, several studies have used the property of circRNAs to be resistant to RNase R, an exonuclease that specifically degrades linear RNAs. Therefore, the use of libraries depleted in the majority RNAs (rRNAs and mRNAs) and treated with RNase R allowed the enrichment in circRNAs and thus eased their identification. As a result of these strategies, hundreds of circRNAs were identified and characterized (22) (circBase, http://www.circbase.org).
However, even if these approaches indeed allowed significant enrichment in circRNAs, a large proportion of linear RNAs escapes the digestion by RNase R. In the recent study by Panda et al. that we highlight here (Figure 2), an additional step has been added to overcome this issue and to remove the remaining RNase R-resistant linear RNAs (1). The strategy was to perform a poly(A) tailing reaction on the remaining linear RNAs, i.e., with a free 3’-OH end, such as those that are processed in a posttranscriptional manner and inherently lack a poly(A) tail (miRNAs, snoRNAs, snRNAs, etc.), or that escape digestion because of their highly structured folding. A poly(A) depletion is then perform on the modified RNA samples. This strategy, called “RNase R treatment followed by polyadenylation and poly(A)+ RNA depletion” (RPAD, Figure 2), was used to isolate highly enriched circRNAs from total RNA (1). However, it should be noted that this approach also removes ciRNAs since they still have a 3’-OH extremity, until trimming of the lariat is complete, that is free for the re-polyadenylation procedure (Figure 1).
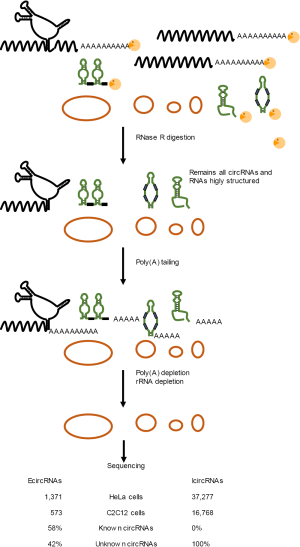
With this new strategy, the authors were able to identify a large number of circRNAs in human HeLa and murine C2C12 cells (38,651 and 17,341 respectively) and release their full-length sequences. Of these, 1,374 were identified as EcircRNAs generated from back-splicing of exons, including 783 already known EcircRNAs (57%), which validated their method. In addition, they also identified 591 new EcircRNAs (43%). Maybe more striking, they uncovered high numbers of yet unidentified IcircRNAs (Figure 1) (37,277 in human and 16,768 in mouse). The authors further experimentally validated some EcircRNAs and IcircRNA candidates by reverse transcription-polymerase chain reaction (RT-PCR) using divergent primers and confirmed the backsplice consensus signature by sequencing. In agreement with the idea that circRNAs may function as sponges for RBPs, the authors also identified several binding sites for RBPs in silico (8,14), supporting the implication of circRNAs in molecular circuitries.
Against all odds, because circRNAs are defined to originate from splicing reactions, some of the newly identified circRNAs mapped to intergenic regions. However, this is an interesting finding that may simply imply that the host transcript of these circRNAs has not yet been identified and annotated in the chosen cellular contexts.
As mentioned above, Panda et al. identified for the first time IcircRNAs whose biogenesis still remains unclear (1). The biogenesis of IcircRNAs might be different form that of EcircRNAs, i.e., through back-splicing. However, they are not produced by canonical splicing like ciRNAs, which were eliminated from the analysis (see above). Despite their abundance revealed by this study, IcircRNAs may originate from poorly preserved splice junctions that cannot be used as a predictive signature to discover additional IcircRNAs.
Again, it is worth noting that IcircRNAs were 20-fold more abundant than EcircRNAs. The most reasonable explanation comes from the genome composition; indeed, introns represent about half the human genome, whereas exons account for only 2% of it (23). Despite the difficulty of studying IcircRNAs as suggested by the authors—e.g., IcircRNAs do not share a well-defined consensus backsplice junction—and given their abundance in the cell, their characterization and mechanisms of action remains to be quickly clarified.
Conclusions
Panda et al. have developed a new method, called RPAD, to enrich circRNAs. With this strategy, they identified thousands of known and unknown circRNAs. More importantly, they discovered a new class of abundant IcircRNAs. We are convinced that this new method is an important first step towards the characterization and the comprehension of the involvement of circRNAs in cells.
Acknowledgments
Funding: Our study was supported by AFM, Association Française contre les Myopathies, Research Grant (#20534) and AFM PhD Fellowships (#21363).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Dr. Jin Li (Cardiac Regeneration and Ageing Lab, School of Life Sciences, Shanghai University, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.01.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Panda AC, De S, Grammatikakis I, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 2017;45:e116 [Crossref] [PubMed]
- Arnberg AC, Van Ommen GJ, Grivell LA, et al. Some yeast mitochondrial RNAs are circular. Cell 1980;19:313-9. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993;73:1019-30. [Crossref] [PubMed]
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93:6536-41. [Crossref] [PubMed]
- Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA 1996;2:603-10. [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679-92. [Crossref] [PubMed]
- Cheng J, Zhang Y, Li Z, et al. A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci China Life Sci 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Lee SR, Lykke-Andersen J. Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol 2013;23:504-10. [Crossref] [PubMed]
- Schneider T, Hung LH, Schreiner S, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep 2016;6:31313. [Crossref] [PubMed]
- Schneider T, Bindereif A. Circular RNAs: Coding or noncoding? Cell Res 2017;27:724-5. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Ulveling D, Francastel C, Hubé F. When one is better than two: RNA with dual functions. Biochimie 2011;93:633-44. [Crossref] [PubMed]
- Xu ZQ, Yang MG, Liu HJ, et al. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 2017;8:1149. [Crossref] [PubMed]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. [Crossref] [PubMed]
- Hubé F, Francastel C. Mammalian introns: when the junk generates molecular diversity. Int J Mol Sci 2015;16:4429-52. [Crossref] [PubMed]
Cite this article as: Bogard B, Francastel C, Hubé F. A new method for the identification of thousands of circular RNAs. Non-coding RNA Investig 2018;2:5.


