
Multi-leveled suppression of p53 function by HuR lncRNPs
The advent of next-generation sequencing has revealed that the majority of expressed transcripts do not encode proteins (1). Many of them belong to a large and growing family of long noncoding RNAs (lncRNAs), generally defined as transcripts larger than 200 nucleotides, typically transcribed by RNA polymerase II, spliced, and polyadenylated, but not translated into protein (2). Mounting evidence indicates that lncRNAs are vital components of gene regulation programs in a range of physiological and pathological processes, including the immune response, neuronal function, cardiovascular disease, and cancer (3-6). By interacting with molecular partners such as DNA, RNA, and protein, lncRNAs form scaffolds, guides, double-stranded nucleic acids, or decoys that influence molecular partners and thereby affect specific gene regulatory processes such as transcription, RNA turnover and transport, mRNA translation, and protein stability (7,8). Among this diverse family, several lncRNAs have been reported to be dysregulated in cancer. For example, lncRNAs MALAT1 and HOTAIR function as oncogenes, promoting cell growth and metastasis, whereas lncRNAs GAS5 and MEG3 function as tumor suppressors, inhibiting the proliferation of cancer cells (3,9).
Many RNA-binding proteins (RBPs) have been identified as forming lncRNA-RBP (lncRNP) complexes that enable lncRNA functions at the cellular and organismal levels. Among them, the RBP HuR [also known as embryonic lethal abnormal vision-like 1 (ELAVL1)], binds many lncRNAs and modulates lncRNA actions (10-12). HuR is abundant and ubiquitously expressed in different cell types, participating in diverse cellular functions including proliferation, differentiation, senescence, and the stress response (13,14). Accordingly, HuR has been implicated in controlling myogenesis, immunological responses, integrity of the gastrointestinal tract, and cancer (14-17). HuR was found highly expressed in a wide range of cancers, had a pro-oncogenic impact, and implemented a pro-cancer gene expression program (18). Until recently, this pro-cancer program was believed to be orchestrated through the impact of HuR on target mRNAs, typically by stabilizing and modulating their translation rates (8); in turn, the encoded proteins promoted cell proliferation, angiogenesis and metastasis, and facilitated cancer cell escape from senescence and immune surveillance (14). However, two recent articles have uncovered two HuR target lncRNAs through which HuR represses, in complementary ways, one of the most important cancer pathways: the p53 tumor suppressor program.
TP53 (‘tumor protein p53’) is one of the most frequently mutated genes in cancer. It encodes p53, a transcription factor that has a strong tumor suppressor function. By binding to p53-response elements on regulatory regions of target genes, p53 transcriptionally induces the production of proteins that reduce cell proliferation, trigger apoptosis, and mount the DNA damage response. Besides transcribing coding genes, p53 also induces the transcription of noncoding RNA, including several lncRNAs that participate in the p53 network and thus control cancer progression (19). For example, p53 was shown to induce transcriptionally lincRNA-p21 in response to DNA damage, and lincRNA-p21 in turn bound p53 as co-activator to enhance p53 transcription (20). Similarly, lincRNA-p21 bound hnRNPK as co-activator to implement a transcriptional repression program that is also a hallmark of p53 and includes shutting off the production of anti-apoptotic and proliferative proteins (21). As reviewed recently (19), another p53-induced lncRNA, PINT, bound the Polycomb repressive complex 2 (PRC2) and enabled PRC2 targeting of specific genes for H3K27 tri-methylation and repression. Following transcriptional activation by p53, lncRNA DINO was found to associate with and stabilize p53, establishing a positive feedback loop that further enhanced p53 function. LncRNA PANDA was transcriptionally activated by p53 from the CDKN1A (p21) locus and bound nuclear transcription factor NF-YA, blocking its function and thereby suppressing apoptosis; however, it also was found to bind and stabilize p53, creating another positive feedback loop. Other lncRNAs that are p53 targets, including PR-lncRNA1, PR-LncRNA10, TUG1, Linc-Ror, and PVT1 were found to affect other cancer traits, as reviewed recently (19).
In a recent issue of Cell Reports, Li et al. (22) identified and functionally characterized the p53-regulated lncRNA PURPL in colorectal cancer. PURPL was strongly induced by DNA damage and was predominant in nuclear. Knockout of PURPL by CRISPR-Cas9 in colon cancer cells reduced survival and accelerated apoptosis. Interestingly, microarray analysis of genes differentially expressed in PURPL WT and PURPL KO cells revealed that many p53 target mRNAs were elevated after ablating PURPL, suggesting that PURPL repressed p53. In addition, loss of PURPL increased p53 stability and steady-state levels in colon cancer cells, but only in cells harboring WT p53 (not mutant p53). These findings are particularly relevant considering current anti-cancer efforts aimed at activating p53; for example, ongoing clinical trials employ inhibitors of MDM2 (a protein that triggers p53 ubiquitination and proteolysis) to promote p53 signaling in the absence of DNA damage, and block cancer progression. Li et al. found that loss of PURPL strongly upregulated p53 abundance and dramatically impaired tumor cell growth in vitro and in vivo, collectively indicating that PURPL promoted survival signaling by controlling p53 levels. In this regard, the negative feedback control of p53 levels exerted by MDM2 was recapitulated by lncRNAs such as linc-RoR and as explained below, also by PURPL.
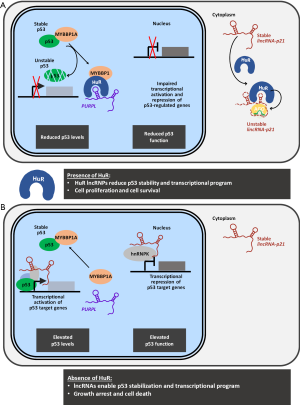
By RNA pulldown and mass spectrometry, Li et al. (22) found strong interaction between PURPL and MYBBP1A, a protein that stabilizes p53. In light of the authors’ result that loss of PURPL increased p53 levels, they proposed that loss of PURPL freed up MYBBP1A, enabling it to bind and increase p53 stability. Interestingly, MYBBP1A is not a canonical RBP and did not bind PURPL; therefore, the authors examined other PURPL-binding proteins from their mass spectrometry survey and identified HuR as a PURPL-interacting RBP. Moreover, they found that MYBBP1A bound PURPL through the adaptor protein HuR, supporting the notion that multiprotein complexes interacting with a lncRNA critically influence its biological function. This paradigm further underscored the notion that a lncRNA can serve as a platform to support the physical interaction of different proteins to elicit a biological outcome. Importantly, in the presence of PURPL, the interaction of HuR with MYBBP1A prevented MYBBP1A from binding to and stabilizing p53, thereby suppressing p53 function.
As mentioned above, HuR was found to bind and regulate the function of several cancer-associated lncRNAs, including OIP5-AS1, MALAT1, and HOTAIR. However, HuR binding to lincRNA-p21 is particularly relevant to the regulatory paradigm described for PURPL. As shown by Yoon et al. (10), the interaction of HuR with lincRNA-p21 led to the recruitment of the microRNA let-7 and associated RISC (RNA-induced silencing complex) to lincRNA-p21, resulting in lincRNA-p21 decay; in turn, the HuR-mediated loss of lincRNA-p21 rescued the translational repression of mRNAs partially complementary with lincRNA-p21 (e.g., JUNB and CTNNB1 mRNAs) (10). Importantly, however, the degradation of lincRNA-p21 also precludes its function as cofactor of hnRNPK. Thus, in the absence of lincRNA-p21, hnRNPK is no longer capable of suppressing the transcription of anti-apoptotic and proliferative genes, while the transcription of some p53-induced genes is impaired (20,21), and thus a pro-survival, proliferative phenotype ensues.
To summarize, HuR can suppress p53 function in two complementary ways, both involving HuR lncRNPs. By binding to and degrading cytosolic lincRNA-p21, HuR can impair the transcriptional program of p53 (including the transcriptional repression program carried out by hnRNPK) (10,20,21). By binding to the nuclear lncRNA PURPL, HuR mediates a loss in p53 levels via a reduction in p53 stability. The joint control of p53 levels and function by lncRNPs is depicted in the presence and absence of HuR (Figure 1).
In closing, we still need far greater knowledge of the abundance of PURPL, lincRNA-p21, and the complexes they form with HuR in cancer and non-cancer tissues. However, the studies by Yoon (10) and Li (22) discussed here bring into focus HuR as an RBP that can jointly suppress p53 function and reduce p53 levels. With escalating interest in developing therapies that raise p53 activity, particularly in combination with DNA-damaging treatments, new lines of effort could be devised based on these findings. For example, new strategies could be created to lower HuR levels or function using small chemical inhibitors (some of which have already been reported in vitro) (23,24). Other approaches could seek to reduce PURPL levels using antisense oligomers or other sequence-specific inhibitors. Yet other interventions could be devised to block the interaction of HuR with these lncRNAs, for instance by employing ‘decoy’ RNAs that block the RNA-binding function of HuR or ‘masking’ oligomers that create regions of partial complementarity on lincRNA-p21 and PURPL that render them refractory to HuR binding. As it is often the case, it might be even more promising to devise these approaches in a multi-pronged fashion: eliciting DNA damage while enhancing the p53 response by jointly suppressing HuR function and its association with lncRNAs that thwart p53 signaling.
Acknowledgments
Funding: This work was funded in its entirety by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Jinzhe Zhou (Department of General Surgery, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.01.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Greco S, Gorospe M, Martelli F. Noncoding RNA in age-related cardiovascular diseases. J Mol Cell Cardiol 2015;83:142-55. [Crossref] [PubMed]
- Mowel WK, Kotzin JJ, McCright SJ, et al. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol 2018;39:55-69. [Crossref] [PubMed]
- Idda ML, Munk R, Abdelmohsen K, et al. Noncoding RNAs in Alzheimer’s Disease. Wiley Interdiscip Rev RNA 2018; [Epub ahead of print]. [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013;425:3723-30. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012;47:648-55. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Kang MJ, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res 2014;42:10099-111. [Crossref] [PubMed]
- Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA 2017;8. [PubMed]
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 2010;1:214-29. [Crossref] [PubMed]
- Xiao L, Wang JY. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol 2014;19:46-53. [Crossref] [PubMed]
- Srikantan S, Gorospe M. HuR function in disease. Front Biosci 2012;17:189-205. (Landmark Ed). [Crossref] [PubMed]
- von Roretz C, Beauchamp P, Di Marco S, et al. HuR and myogenesis: being in the right place at the right time. Biochim Biophys Acta 2011;1813:1663-7.
- López de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003;22:7146-54. [Crossref] [PubMed]
- Chaudhary R, Lal A. Long noncoding RNAs in the p53 network. Wiley Interdiscip Rev RNA 2017;8. [PubMed]
- Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014;54:777-90. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Li XL, Subramanian M, Jones MF, et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep 2017;20:2408-23. [Crossref] [PubMed]
- Muralidharan R, Mehta M, Ahmed R, et al. HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci Rep 2017;7:9694. [Crossref] [PubMed]
- Lal P, Cerofolini L, D’Agostino VG, et al. Regulation of HuR structure and function by dihydrotanshinone-I. Nucleic Acids Res 2017;45:9514-27. [Crossref] [PubMed]
Cite this article as: Yang JH, Gorospe M. Multi-leveled suppression of p53 function by HuR lncRNPs. Non-coding RNA Investig 2018;2:2.


