
Can long non-coding RNAs reveal the hidden code of lung cancer?
At present, lung cancer remains the first cause of cancer-related death worldwide, and non-small cell lung cancer (NSCLC) accounts for 80% of cases. Despite advances in the treatment of NSCLC, patient prognosis remains dismal, with overall survival rates ranging from 55% in local disease to 4% in metastatic disease. This scenario, combined with the absence of proper screening tools, highlights the urgency of developing new diagnostic biomarkers that can identify lung cancer patients at early-stage where surgery with curative intent is the preferred treatment. Although early diagnosis is related to increased survival, in the first five years after surgery, up to 40% of resected patients relapse (1). Therefore, it also urges the development of new biomarkers to identify lung cancer patients with a high risk of relapse after surgery in order to improve their clinical management and survival.
In this line, the study of long non-coding RNAs (lncRNAs) has emerged as one of the most promising fields in the development of new biomarkers for all cancer types, including NSCLC (2-4). LncRNAs display an extremely specific expression pattern, and their capacity to discriminate between tumor and normal tissue is superior to coding genes or pseudogenes, making them an ideal source of biomarkers (5).
The recently published manuscript by Acha-Sagredo et al. reports their study of deregulated lncRNAs associated with NSCLC pathogenesis (6) and provides some additional information to the state of the art (2-4). This was two-phase study (Figure 1). An initial discovery data set based on microarray analysis of 44 paired normal and NSCLC tissue was used for the identification of dysregulated lncRNAs in NSCLC. This discovery phase was followed by qPCR validation in 38 additional specimens, which was cross-validated using available TCGA data [n=850: lung adenocarcinoma (LUAD) =585; squamous cell carcinoma (LUSC) =504]. During the discovery phase, a total of 1122 downregulated and 771 upregulated lncRNAs [including long intergenic non-coding RNAs (lincRNAs) and anti-sense transcripts] were identified. The validation phase confirmed the upregulation of six lncRNAs and the downregulation of six other lncRNAs in tumor tissue compared to normal tissue.
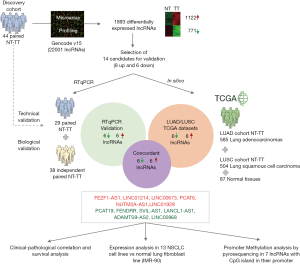
Some of the upregulated lncRNAs identified have previously been described as oncogenes in NSCLC. For example, LINC00673 and PCAT6 have been linked to several key processes in cancer progression: LINC00673 to epithelial-to-mesenchymal transition (EMT) and cellular senescence (7,8); and PCAT6 to the recruiting of chromatin-modifying complexes (9). Consistent with these results, some of the downregulated lncRNAs identified in the study by Acha-Sagredo et al. (6) have previously been described by Salavaty et al. using TCGA LUAD data (10). For instance, they explored and confirmed the role of the lncRNAs LANCL1-AS1, FENDRR, LINC00968 and ADAMTS9-AS2 as tumor suppressors in NSCLC adenocarcinoma. Salavaty et al. analyzed the TCGA data using two different web-servers, circlncRNAnet (http://app.cgu.edu.tw/circlnc/) and GEPIA (http://gepia.cancer-pku.cn/), and studied the lncRNAs that were differentially expressed on both servers. Interestingly, they also used data from plasma RNA-Seq to study circulating lncRNAs and identified SNHG6 as a candidate for further evaluation as a circulating biomarker of lung adenocarcinoma.
While Acha-Sagredo et al. found that PCAT19 was downregulated in tumor tissue, Zhang et al. (11) reported that it was upregulated in NSCLC tumor tissue samples and negatively correlated with the mRNA expression of the TP53 gene in both normal and tumor tissue. They also found a correlation between high expression of PCAT19 in tumor tissue and shorter overall survival (11), but this impact on survival was not observed by Acha-Sagredo et al. Similarly, Acha-Sagredo et al. found that LINC00968 was downregulated in tumor tissue, but it has previously been reported as upregulated in tumor tissue, where it acted as an oncogene (12,13). The authors suggested that the contradictory results observed in these two lncRNAs, PCAT19 and LINC00968, could be explained by differences between databases annotations producing that different transcripts are annotated under the same name.
Acha-Sagredo et al. further explored the differential expression of the dysregulated lncRNAs in normal and tumor tissue by comparing their expression in a panel of 13 NSCLC cell lines and a normal lung embryonic fibroblast cell line (IMR-90). In our opinion, the normal cell line used for this comparison was not the most adequate since fibroblasts have a different embryonic origin (mesoderm) than the epithelial cells (endoderm) that trigger NSCLC. Probably, the selection of a normal lung epithelial cell line, such as BEAS-2B or other, would have been a better control for this comparison. Furthermore, functional studies in these cell lines to unravel the role in the carcinogenesis process of the lncRNAs identified is missing.
The authors also analyzed the correlation between the expression of the lncRNAs and patient clinicopathological characteristics in both their cohort and in the TCGA dataset (Figure 2). In their cohort they found correlations between sex and the lncRNAs FEZF1-AS1 and ADAMTS9-AS2 and between tumor differentiation grade and the lncRNAs PCAT6 and LANCL1-AS1. However, probably, one of the most salient findings in this line is the correlation between the presence of lymph node metastasis and the lncRNAs LINC01929 and LINC00968. Lymph node status (N) is a significant component of staging of NSCLC and is directly related to relapse rate after surgery and with treatment decisions. Patients are classified depending on the location of the metastatic lymph node as N0 (no nodal involvement), N1 (peribronchial, interlobar, hilar node involvement), N2 (ipsilateral nodal involvement) or N3 (contralateral mediastinal, contralateral hilar or supraclavicular nodal involvement (14). Although the authors not deepened in the study of lncRNAs and the N type, the identification of lncRNAs linked to N status is for sure of clinical importance. For instance, N2 occult lymph node involvement is present in 10% of stage I NSCLC patients and in 21% of those with stage II (15), which can impact the clinical management of patients. Although stage I patients are not usually treated with adjuvant chemotherapy, if N2 involvement is confirmed, they may increase to stage IIIA, where adjuvant treatment has demonstrated benefit in outcome (1,14). To our knowledge, the identified association between N and the lncRNAs LINC01929 and LINC00968 have not been previously described in NSCLC, but the authors were not able to crossvalidate this finding in the TCGA cohort. In fact, in the TCGA analysis other lncRNAs from the validation list were significantly correlated with N, including LINC00673, LANCL1-AS1 and ADAMTS9-AS2, and LINC00968 was found with and opposite relationship than in their cohort. Two of the lncRNAs associated with N in the TCGA dataset have been previously described in the literature. LINC00673, which was upregulated in patients with lymph node involvement, has been also related to lymph node metastasis in gastric, thyroid, breast and ovarian cancer (16-19). Furthermore, ADAMTS9-AS2, which was downregulated in patients with lymph node involvement, has also been reported to be downregulated in ovarian and gastric cancer (20,21). Despite the differences between the discovery and validation cohort in this matter, the role of these lncRNAs in lymph node involvement merits further exploration and with further research and validation, they might become a powerful tool for clinical practice.
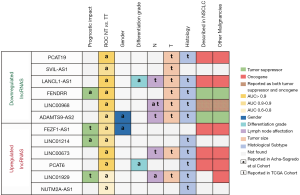
Finally, the authors point out that some of the studied lncRNA could become deregulated by alterations in the methylation patterns of the CpG islands located in their promoters. Specifically, the methylation status of 7 out of the 14 lncRNAs was analyzed: PCAT6, RNF139-AS1, LINC00673 and NUTM2A-AS1. Although the results were not conclusive since no correlation between expression levels and degree of methylation was observed in the paired samples, the epigenetic mechanism of lncRNA dysregulation is a point that deserves further study.
Research on lncRNAs needs to deal with several concerns to reinforce their clinical applicability. The authors denoted that one of the most challenging concerns in lncRNA research is the lack of proper annotation for most of these genes and the differences in sequence and genomic location between different databases. It is urgent, therefore, to increase efforts towards a global consensus on these issues in order to improve the replicability of the results and their potential clinical application (22). Moreover, recently, the study of biomarkers that can be screened in body fluids (liquid biopsy) has emerged as a key direction for translational research. Several studies have explored the role of circulating lncRNAs as diagnostic biomarkers in NSCLC (23). Additionally, others, including our group, have highlighted the synergistic advantages of exploring the role of lncRNAs in extracellular vesicles as part of the liquid biopsy (24). Furthermore, new, clinically relevant sources of lncRNAs have emerged, such as the use of the tumor-draining pulmonary vein, which can be accessed during surgery and has proven to be enriched in primary tumor-secreted lncRNAs (25).
In conclusion, the present study reinforces the idea that lncRNAs are new promising biomarkers that might help us to shed light on the pathological mechanisms of NSCLC tumorigenesis and progression, considering in our humble opinion, the new hidden code of NSCLC.
Acknowledgments
Funding: This work was supported by a grant from the Ministry of Economy, Industry and Competition, Agencia Estatal de Investigación co-financed with the European Union FEDER funds SAF2017-88606-P (AEI/FEDER, UE).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Non-coding RNA Investigation. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form and declare (available at http://dx.doi.org/10.21037/ncri.2020.03.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vansteenkiste J, Crinò L, Dooms C, et al. 2nd ESMO Consensus Conference on Lung Cancer: early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann Oncol 2014;25:1462-74. [Crossref] [PubMed]
- Zhou W, Liu T, Saren G, et al. Comprehensive analysis of differentially expressed long non-coding RNAs in non-small cell lung cancer. Oncol Lett 2019;18:1145-56. [PubMed]
- White NM, Cabanski CR, Silva-Fisher JM, et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 2014;15:429. [Crossref] [PubMed]
- Yang J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer 2014;85:110-5. [Crossref] [PubMed]
- Yan X, Hu Z, Feng Y, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 2015;28:529-40. [Crossref] [PubMed]
- Acha-Sagredo A, Uko B, Pantazi P, et al. Long non-coding RNA dysregulation is a frequent event in non-small cell lung carcinoma pathogenesis. Br J Cancer 2020;1-9. [Epub ahead of print]. [PubMed]
- Kim DU, San Lucas FA, Cagley M, et al. Larger size extracellular vesicle as early biomarker in patients with metastatic pancreatic cancer. J Clin Oncol 2017;35:234. [Crossref]
- Roth A, Boulay K, Groß M, et al. Targeting LINC00673 expression triggers cellular senescence in lung cancer. RNA Biol 2018;15:1499-511. [Crossref] [PubMed]
- Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine 2018;37:177-87. [Crossref] [PubMed]
- Salavaty A, Rezvani Z, Najafi A. Survival analysis and functional annotation of long non-coding RNAs in lung adenocarcinoma. J Cell Mol Med 2019;23:5600-17. [Crossref] [PubMed]
- Zhang X, Wang Q, Xu Y, et al. lncRNA PCAT19 negatively regulates p53 in non-small cell lung cancer. Oncol Lett 2019;18:6795-800. [PubMed]
- Wang Y, Zhou J, Xu YJ, et al. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J Cell Physiol 2018;233:3397-406. [Crossref] [PubMed]
- Xiong Y, Zhang X, Lin Z, et al. SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1, and FEZF1-AS1 are crucial long non-coding RNAs associated with the prognosis of lung squamous cell carcinoma. Oncol Lett 2019;18:3985-93. [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Dyas AR, King RW, Ghanim AF, et al. Clinical Misstagings and Risk Factors of Occult Nodal Disease in Non-Small Cell Lung Cancer. Ann Thorac Surg 2018;106:1492-8. [Crossref] [PubMed]
- Ba MC, Long H, Cui SZ, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget 2017;8:95542. [Crossref] [PubMed]
- Xia E, Bhandari A, Shen Y, et al. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int J Oncol 2018;53:1927-38. [PubMed]
- Xia E, Shen Y, Bhandari A, et al. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res 2018;8:1273. [PubMed]
- Zheng T, Qiu J, Li C, et al. Long noncoding RNA LINC00673 promotes the proliferation and metastasis of epithelial ovarian cancer by associating with opioid growth factor receptor. Onco Targets Ther 2019;12:6145. [Crossref] [PubMed]
- Wang A, Jin C, Li H, et al. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int J Biol Macromol 2018;120:1705-13. [Crossref] [PubMed]
- Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother 2018;107:185-93. [Crossref] [PubMed]
- Weirick T, John D, Uchida S. Resolving the problem of multiple accessions of the same transcript deposited across various public databases. Brief Bioinform 2017;18:226-35. [PubMed]
- Hu X, Bao J, Wang Z, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol 2016;37:3497-504. [Crossref] [PubMed]
- Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res 2018;7:S243-52. [Crossref] [PubMed]
- Castellano JJ, Marrades RM, Molins L, et al. Extracellular Vesicle lincRNA-p21 Expression in Tumor-Draining Pulmonary Vein Defines Prognosis in NSCLC and Modulates Endothelial Cell Behavior. Cancers 2020;12:734. [Crossref] [PubMed]
Cite this article as: Castellano JJ, Navarro A. Can long non-coding RNAs reveal the hidden code of lung cancer? Non-coding RNA Investig 2020;4:6.


