
Regulatory roles of small RNAs in prokaryotes: parallels and contrast with eukaryotic miRNA
The capacity of RNA to store information, transfer information, and perform catalysis makes it one of the most functionally diverse molecules in biology. In addition to the well characterized role of mRNA, tRNA, and rRNA, RNA molecules have now been identified that have regulatory roles in the transcription and translation of mRNA. The regulatory roles of RNA are best characterized of micro RNAs (miRNAs) in eukaryotes that act to suppress mRNA translation by complimentary binding of the miRNA to the mRNA, inhibiting the mRNA translation by the ribosome, and in some cases, signaling the degradation of the mRNA (1). While the first instance of a regulatory RNA in a prokaryote was identified over fifty years ago, the identification and characterization of most regulatory RNAs in prokaryotes have only been identified this century (2-5). With the development of methods in transcriptomics and RNA sequencing technology, the number of regulatory RNAs and potential regulatory RNAs in prokaryotes has grown rapidly, and several RNA regulated systems and cell circuits have been characterized (2,3,5-12).
The most common description of regulatory RNAs are non-coding, ncRNAs, that are completely or partially complementary to a target RNA molecule and inhibit or activate the transcription or translation of one more mRNAs by Watson-Crick base pairing between the regulatory RNA and the target molecule (2,3,5). Currently, the most researched target RNA molecule is mRNA, but recent reports have also identified regulatory ncRNAs that target rRNA as well (13). While the mechanism of RNA inhibition by transcriptional interference is considered an RNA regulatory process, it occurs by a mechanism that is unrelated to the complementary binding between the target and the regulatory RNA (14). All other described mechanisms of regulation by RNAs including transcription attenuation (15), translational inhibition or activation (7,9,16), and mRNA protection (17,18) all require Watson-Crick base pairing between the target and the regulatory RNA (discussed below).
RNA terminology
Increased interest in regulatory RNAs and the advancements in RNA sequencing technology has allowed a rapid increase in the identification of regulatory RNAs from various sources and that function by various mechanisms. The increase in research of regulatory RNA has in many instances outpaced the rate of the terminology, and a review of the literature reveals a lack of consistency in the use of naming conventions for various types of RNAs. As noted by various research groups this lack of consistency can be largely attributed to advancements in our understanding of the roles of regulatory RNAs, and a lack of understanding of how to classify RNAs due to incomplete information of the relationship between origins, size, sequence, structure, function and mechanism of regulatory RNAs (5). Therefore, the most consistent methods for classifying and naming different regulatory RNAs are related to the target of the regulatory RNA, the location of the DNA coding sequence of the regulatory RNA in relation to the operon of the mRNA target, and the size of the regulatory RNA. In many instances, the name of the regulatory RNA is dependent on the context in which the regulatory RNA is being discussed, which occurs mainly for RNAs for which new functions have been discovered. For example, the term non-coding RNA has traditionally been used for RNA that are not translated into protein. However, recent sequencing results have shown that many of these ncRNAs contain ORFs and can be translated into protein or small peptide (5). Additionally, it has been suggested that some mRNAs also act as regulatory RNAs in some instances. It is likely that the naming scheme of regulatory RNAs will evolve as the research advances to allow the classification of these RNAs based on the above-mentioned characteristics, but for now, the convention is that each researcher defines the terms as they will use them in there reporting. In this review, we have outlined the names and descriptions of the RNAs to be discussed in Table 1.
Table 1
| Name of RNA | Abbreviation | Description |
|---|---|---|
| Non-coding RNA | ncRNA | Any RNA that is not mRNA. Because it has now been reported that some RNA molecules originally thought to only act as non-coding RNA have been shown to be translated into small peptides, the term ncRNA is used here in the context of the function of the RNA at that time |
| Trans-encoded RNA | trans | A regulatory non-coding RNA that is transcribed DNA that lies outside the operon of the target mRNA |
| Cis-encoded RNA | cis | A regulatory non-coding RNA that is transcribed from within the same operon as the target mRNA |
| Trans-acting non-coding RNA | None | A regulatory non-coding RNA that is transcribed as a separate RNA strand from the target mRNA |
| Cis-acting RNA | None | A regulatory non-coding RNA that is transcribed in the same strand as the target mRNA, typically in the 5’ UTR |
| Long non-coding RNA | lncRNA | Any ncRNA that is greater than ~200 nucleotides |
| Short non-coding RNA | sncRNA | A ncRNA that is shorter than ~100 nucleotides long |
| Micro-RNA | miRNA | A 22 nucleotide trans-encoded, trans-acting, non-coding RNA that regulates the transcription of mRNA in eukaryotes. miRNA is originally transcribed as lncRNA, pri-miRNA, and is processed first to a sncRNA, pre-miRNA by the RNase, Drosha, and the processed again to a duplex miRNA by the RNase, Dicer. One of the strands in the duplex miRNA is partially complementary to the target mRNA and suppresses the mRNA translation by Watson-Crick bonding to the mRNA which is modulated by Argonaute protein |
| Small interfering RNA | siRNA | Functions by the same mechanism as miRNA, but the length can vary slightly from 22 nucleotide. siRNA is also trans-encoded, trans-acting, non-coding RNA, but with different origins than miRNA, including transfection. The initial transcript is processed by the Dicer as with miRNA, and Argonaute protein is needed to facilitate binding |
| Small RNA | sRNA | Trans-acting, trans-encoded regulatory sncRNA. sRNA is only partially complementary to the sequence of the mRNA regulated. Silencing occurs by binding the mRNA through Watson-Crick base pairs and requires a chaperone protein, typically Hfq, to bind the regulated mRNA |
| Antisense RNA | asRNA | Trans-acting cis-encoded regulatory sncRNA. asRNAs are completely or mostly complementary to the sequence of the mRNA regulated. Silencing occurs by binding the mRNA through Watson-Crick base pairs with > 75 nt complimentary overlap between asRNA and target mRNA. asRNA do not typically requires a chaperone protein, to bind the regulated mRNA |
| Riboswitches | None | Cis-acting, cis-encoded regulatory RNAs. Typically located in the 5’UTR of the mRNA that they regulate. Regulate translation and transcription of the mRNA by adopting different secondary structures in response to various substrates |
| Ribosome associated non-coding RNA | rancRNA | Trans-acting, trans-encoded lncRNA that regulates translation by through interactions with the ribosome |
The regulatory RNAs discussed here are small regulatory RNAs (sreRNAs) classified as antisense RNAs (asRNAs) short RNAs (sRNAs) and micro-like size RNA (msRNA) and their regulatory roles in prokaryotes. Both asRNAs and sRNA act as trans-regulatory elements (TREs) to their target RNAs, in that both regulatory RNAs are initially separate molecules from their targets, typically mRNA but occasionally a rRNA, a tRNA, or another regulatory RNA. Here, asRNAs are strictly defined as RNAs that are cis-encoded, coded on the DNA within the same genetic loci as the target RNA but on the opposite DNA antisense strand as the target RNA (3). sRNAs are coded in a DNA sequence outside of the genetic loci of the target. Due to the difference in the origins of the two regulatory RNAs, the asRNAs are completely or almost completely complementary to their target RNAs, while sRNAs are only partially complementary to their target RNAs. The term asRNA will often be used in a way that it includes sRNA due to similarities in the Watson-Crick base pairing interaction mechanism shared by both RNA. However, due to the difference in the origins of the molecules and the difference in complementarity of the regulatory RNAs with their targets, complexes of asRNAs and sRNAs with their targets may result in different levels of regulation, different downstream processes, and often involve different proteins in the mechanisms of regulation (2,3,6,7). Therefore, they are kept as separate classes of regulatory RNAs here, and the term sreRNAs will be used when referring to both classes of RNAs.
Cellular functions regulated by sreRNAs
The mechanism of regulation of transcription and translation by sreRNAs is typically by Watson-Crick base pairing between the regulatory RNA and the RNA target. The process that is regulated, transcription versus translation, is dependent on the target RNA, the region of binding on the RNA, environmental factors present, and secondary structural elements present or created upon binding (2,3,6,9). Transcriptional attenuation has previously been shown to occur when the binding of a small molecule, ribosome, tRNA, or protein bind the target mRNA during the transcription of the mRNA causing a disruption and termination of the transcription process (15). This mechanism has recently been described in the regulation of tryptophan biosynthesis due to the binding of a sRNA, rnTrpL (19). In this instance, rnTrpL downregulates the expression of the trpDC operon. The expression of trpDC suppresses tryptophan biosynthesis when the levels of tryptophan are low. A stem-loop, SL1, region of rnTrpL base pairs trpD mRNA. The binding of the rnTrpL attenuates the transcription of the trpDC operon.
The silencing of mRNA by sreRNAs by translational inhibition is the most reported and appears the most common regulatory actions of sreRNAs (6,7). Inhibition of translation by regulatory RNAs can also involve the binding of the regulatory RNA to the ribosome binding site on the 5’ UTR of the target mRNA, and thus inhibiting translation by blocking the docking of the mRNA with the ribosome. The mechanism was observed in the silencing of CopT mRNA by the asRNA CopA. The interaction occurs through initial contacts between two loops on each RNA in what is termed a kissing complex. The kissing complex then progresses to form a four-way junction that sequesters the ribosomal binding site. The silencing of CopT inhibits the replication of plasmid R1 in E. coli (20,21). An early example of a sRNA that inhibits transcription is RNAIII. RNAIII is an mRNA encoding delta-hemolysin peptide in S. aureus, and acts as a sRNA of multiple target mRNAs (22,23). RNAIII inhibits the translation of the spa mRNA during exponential growth phase. The binding of the two RNAs occurs in the 3’-domain of RNAII and the 5’UTR of spa mRNA. In both instances, the creation of the dsRNA of the complex signals dsRNA specific RNases to cleave the mRNA.
While inhibition of translation is the most common regulation by RNAs, sreRNAs have also been observed that stimulate translation (24). The stimulation of translation occurs by the disruption of a secondary structure in the mRNA which contains the ribosome binding sequence by the binding sreRNA. The disruption of the secondary sequence exposes the ribosomal binding site and translation proceeds. This mechanism was observed in the binding of RNA III with hla mRNA (24). In addition to its role in mRNA silencing by the binding of spa mRNA, the above mentioned RNAIII in S. aureus also binds the hla mRNA. However, the binding of the hla mRNA disrupts a secondary structure in the hla mRNA and exposes the ribosomal binding domain. Thus, the binding of RNAIII to hla mRNA results in the activation of the translation of the mRNA (24). In a different type of translational stimulation, the gadXW mRNA is initially transcribed as a long unstable transcript. The binding of the GadY asRNA to the 3’ end induces the processing of the gadXW mRNA into a more stable transcript (25).
A final regulatory function observed by the sreRNAs is protection of mRNA by RNases that specifically cleave single stranded RNA. This mechanism has been observed during viral infection of phage P-SSP7 into the cyanobacterium Prochlorococcus MED4. Upon infection the phage transcript inhibits the promoter of the rne gene inhibiting the major form of RNase E. This inhibition of the production of RNase E results in the accumulation of a shorter, more stable form of RNase of RNase E. At the same time the phage produces asRNA that covers most of its genomic transcipts. The phage asRNAs binding to the phage mRNA protects the mRNA from cleavage by the short RNase E, while the RNase then cleaves the single stranded mRNA of the host (17,18). The preservation of the phage mRNAs and the degradation of the host mRNA allows the rapid translation of the phage proteins. Additionally, specific asRNAs are transcribed by the host that protects select mRNAs from degradation. It is suggested that the protected host mRNAs are useful for the phage replication.
Binding of sreRNA to target mRNA
The silencing of the mRNAs by sreRNAs is mediated by Watson-Crick base pairing between the two RNAs. The formation of the silencing complex is dependent on two factors: (I) interaction of a seed sequence; and (II) local concentration of the sreRNA. The seed regions are 5 to 7 nt regions required for initial interactions between the sreRNA and the target mRNA (26). The seed sequences are typically located in exposed loop regions of the sreRNA, the target mRNA, or both. The interactions between the seed sequences between the two RNAs are rate limiting step in the formation of the complex. The efficiency of formation is dependent on the on rate the formation of the initial seed interaction, in which a faster binding rate is favored over a slower more stable interaction (26).
The interactions between the seed regions can occur between loops located on each of the RNAs in what is termed a kissing complex (6,26,27). The kissing complex can occur between one or two loops located on each RNA, and the kissing complex appears to be the most common mechanism for initial ‘seeding’ of the complex. However, the seed sequences can interact between a loop region and a single-strand sequence of the RNAs. For instance, the loop-single strand ‘seeding’ has been observed in the regulation of the hok mRNA by the asRNA Sok interaction. In the Sok/hok system, the loop region is present on the hok mRNA and the 5’ region of the Sok asRNA single-stranded (28).
Following the initial seed interaction, an extensive formation of ds-RNA duplex is propagated between the complimentary regions of the sreRNA and the mRNA. The duplex formation can occur in a single step or multiple steps. The single step duplex formation has been well characterized in the hok/Sok system, and propagation occurs from the initial ‘seed’ in a ‘zipper-like’ mechanism (28). The propagation of the ‘zipper’ is dependent on a ‘bulge’ in the stem of the loop region and imperfect complementarity in the stem that gives way to the more stable structure resulting from greater complementarity between the sreRNA and the mRNA target (6). The multiple step system has been best demonstrated in the CopT/CopA system, in which the multiple step mechanism occurs by multiple intermediate structures between the RNAs (26). In the multiple step mechanism, the initial site of helix propagation occurs at a site that is located separately from the initial seed sequence. The ‘seeding’ interaction brings complementary single strands of each RNA into proximity of each other, resulting in the initial formation of the stable duplex RNA structures.
The regulation of transcription of sreRNAs
The regulation of the transcription of sreRNAs appears to be predominately under the control of σ-factors in a mechanism similar to their mRNA targets (8). However, the level of the sreRNAs is lower than those of the target mRNAs, so it is likely that other factors affect the transcription of sreRNA. Stress related σ factors specific to the sreRNA promoter have also been shown to control the transcription of sreRNAs in response to stress caused by ethanol levels in E. coli (29). Additionally, the presence of a riboswitch in the leader sequences of an asRNA was reported to regulate the transcription of asRNAs in Clostridium acetobutylicum. The riboswitch upstream from the asRNA responds to specific levels of sulfur to regulate transcription of asRNA (30).
The termination of sreRNA is typically achieved by Rho-independent termination (31). Rho-independent termination is achieved by the presence of a GC-rich dyad repeat that forms a stem-loop followed by a T-rich stretch. The T-rich sequence gives rise to a U-rich tail on the resulting sreRNA. The termination sequence of asRNA is structurally conserved in that the formation of a stem-loop is required for the Rho-independent termination, but sequence is not conserved, and is only required to be GC-rich (31). The presence of at least seven Ts is observed for all Hfq interacting sreRNAs (see below). The conservation the Ts in the T-rich sequence likely arises due to the necessity of HFq to interact with at least seven Us for functional binding (31). In addition, the 3’OH that arises from Rho-independent termination is also required for the interaction with Hfq (32). The conservation of the T-rich sequence may help in identifying the coding sequences of sreRNAs in future studies (31).
asRNA vs. sRNAs
In prokaryotic systems, a similar mechanism of translational inhibition has been reported involving asRNAs and sRNAs. Mechanisms of both regulatory RNAs involve the Watson-Crick binding to the target mRNA. However, differences in the origins of the sreRNAs may lead to differences in mechanisms of RNA regulation. As mentioned above, asRNAs bind to their target mRNAs by complete, or almost complete, complementarity of base pairs between the overlapping regions of the two RNA molecules. The extensive complementarity between the asRNA and the target mRNA is due to transcription of the antisense strand of the target mRNA gene referred to as pervasive transcription (3). The location of the complementary overlapping region on each RNA molecule is determined by the relative position of the promoter of the asRNA to the mRNA target within the operon of the DNA. The interaction between the asRNA and mRNA has been described as “head to head” if the overlapping regions are in the 5’ end of each strand, “tail to tail” if the overlapping regions are in the 3’ of each strand, or “fully” if the overlapping region spans the entire asRNA sequence (8,9). Regardless of where the overlapping region occurs, there are >75 canonical base pairs involved in Watson-Crick base pairs involved in the regulatory complex made up from an asRNA strand and a strand from the target mRNA (16). The regulation by asRNA has been most closely linked to transposable elements on plasmids, and phage and toxin genes, but asRNAs regulation has now been observed in responses to different stresses (3,6). Despite reports of interaction with Hfq (see below), association with asRNA and a chaperone protein is not typically required for the interaction with the target mRNA.
The origins of sRNAs can typically be traced to endogenous sources, RNA transcribed from endogenous DNA resulting from transpositions, or exogenous sources, RNA transcribed from exogenous DNA or exogenous RNA resulting from horizontal gene transfer or viral infection. To date, more sRNAs have been identified and appear to play a greater role in RNA regulation compared to asRNAs. sRNAs are more often associated with adaptive stress responses, including antibiotic resistance. sRNAs lack complete complementarity to the target mRNA and have fewer bases in the overlapping region (3,8,33). Additionally, sRNAs typically rely on chaperone proteins to facilitate the interaction with the target mRNA. Therefore, Rho-independent termination sequence is typically required (31).
miRNA-like mechanism in prokaryotes
Translation inhibition by a regulatory RNA has been most extensively characterized in eukaryotic systems by miRNAs (34). In these systems, miRNAs are initially embedded in a long non-coding transcript several hundred nucleotides long referred to as the primary-miRNA, pri-miRNA (Figure 1). The pri-miRNA is processed by the RNase III nuclease, Drosha, in the nucleus of eukaryotes to a ~70 nucleotide pre-miRNA with the guidance of DGCR8. The pre-miRNA is exported to the cytoplasm Exportin-5 where it processed to the mature duplex miRNA by the RNase III enzyme, Dicer into a 22 nucleotide mature duplex miRNA. Following processing by Dicer, a single strand of the mature miRNA that is partially complementary to the target mRNA is loaded onto a chaperone protein, Argonaute (Ago), that facilitates the binding of the miRNA to its target mRNA creating the RISC complex. The resulting dsRNA silences the mRNA translation or signals dsRNA specific RNase to cleave the mRNA. The silencing versus the degradation of the mRNA target depends on the degree of complementarity between the miRNA and the mRNA. In both instances of sequestering or degradation of the mRNA, the translation of the mRNA is inhibited (34).
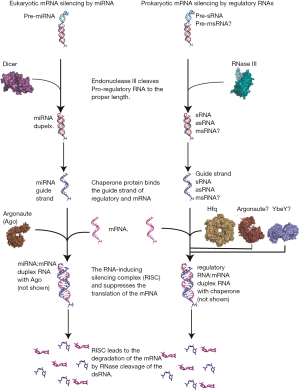
A mechanisms with the characteristics of miRNA or siRNAs in eukaryotes was deemed unlikely in prokaryotes due to the lack of persistence of sreRNAs capable of binding to target mRNAs, and the lack of the additional regulatory machinery necessary to process lncRNAs, similar to primary and pre-miRNA transcripts, or form the RISC-like complex responsible for silencing the translation of mRNAs (35,36). However, a renewed interest in the search of miRNA-like prokaryotic mechanism has risen due to recent advancements in RNA sequencing technology that allowed the identification of persistent msRNAs and the identification of potential chaperone proteins for sRNAs that could potentially perform regulatory roles (10,37).
The search for miRNAs sized RNAs
In addition to identifying potential machinery necessary to perform the miRNA-like regulation, Next Generation RNA sequencing has also identified micro-size RNA molecules, msRNAs. msRNAs are the product of the processing of longer strands of RNA by RNases, the product of disrupted transcription, or pervasive transcription (38-41). Due to small number of nucleotides involved in the ds msRNAs, any regulatory complexes identified involving msRNAs will likely require proteins to facilitate complex formation with the target mRNA in a similar mechanism to miRNAs and siRNAs in eukaryotes (as mentioned above) or sRNAs (see below). The duplex formed by msRNA with the target mRNA are less thermodynamically stable due to the small number of nucleotides involved in the complex. Currently, regulation of mRNAs by msRNAs has only been identified from pathogenic bacteria, and require the host machinery for regulation.
Dicer like RNase
As discussed above, Dicer is an endoribonuclease responsible for the cleavage of the ~70 nt pre-miRNA to the mature duplex 22 nt miRNA in eukaryotes (42). Dicer is an ortholog of the bacterial RNase III and contains two RNase III domains that form an intermolecular dimer that cleaves dsRNA. In addition to the catalytic RNase III domains, Dicer has a PAZ domain responsible for anchoring the 3’ and 5’ ends of the dsRNA, which is used to determine the product size, an ATPase/helicase domain, a domain of unknown function (DUF283) and a dsRNA binding domain. The structure of Dicer gives rise the specific length of miRNAs and the higher rate of cleavage of pre-miRNA compared to pre-siRNAs (43). The majority of prokaryotic regulatory RNAs are reported to silence their target mRNA without processing of the original transcript (41). However, a Dicer like mechanism may play a role in the processing of a pre-regulatory RNA.
The sRNA, ArcZ, is responsible for the suppression of sdaC, STM3216 and tpx mRNAs in Salmonella. The transcript of ArcZ is ~120 nt, however, the pre-ArcZ is processed to an ~50 nt functional sRNA before binding to the target mRNAs (44). Similarly, the sRNA, MicL, which silences the mRNA encoding for the membrane lipoprotein Lpp, is transcribed as a 308 nt pre-sRNA and is processed to an 80 nt functional tsRNA (45). In both instances, the ribonuclease responsible for the processing was not determined. However, in the latter case of MicL, the authors were able to rule out RNase E, RNase III, RNase G, RNase BN, and YbeY as the ribonuclease responsible. The lack of identification of the processing ribonuclease gives rise to the possibility of an uncharacterized RNase present, or multiple RNase targeting the primary transcript.
Although no RNase has been directly linked to the processing of pre-msRNAs to the mature msRNAs, RNase III has been suggested to process primary transcripts of regulatory RNAs to produce RNA of similar size to miRNAs in eukaryotes. E. coli RNase III contains a C-terminal dsRNA binding domain and a N-terminal catalytic domain. The RNase III homodimer cleaves pre-mRNA to the mature mRNA in a mechanism similar to that of Dicer cleavage of miRNA (43). While the lack of a PAZ domain in RNase III gives rise to products of various length, RNA sequencing data of RNAs less than 50 nt in E. coli showed that RNase III correlated processing of asRNA duplexes gave rise to ~20 nt length products in a process that appears to be conserved in Gram positive bacteria (33). In addition to RNase III, the endoribonuclease MazF has been suggested to act as the Dicer-like processor in some instances in stress responses (39). While it is still unclear if any of the msRNAs are functional as regulatory RNAs, it appears that the Dicer like activity exists to create a similar RNA unit.
Chaperone proteins and a RISC-like complex
One of the hallmarks of the regulation of mRNA by miRNAs in eukaryotes is the formation of the RNA-induced silencing complex which is formed by the miRNA guided strand, the mRNA coding strand loaded onto the RNA binding Argonaute protein (Ago) (46,47). The formation of the RISC complex results in the suppression of the translation of the mRNA or the proteolytic cleavage of the mRNA depending on its complementarity with the miRNA (1,48). For asRNA in prokaryotes, the extensive complementarity between the asRNA and the target mRNA appears sufficient for a stable complex between the two RNAs. However, the formation of a stable duplex between sRNAs and mRNAs likely requires the formation of a RISC type complex aided by a chaperone RNA binding protein for formation. The presence of a chaperone would be especially necessary if it is shown that msRNAs do indeed perform a regulatory role in prokaryotes. Several candidates for the role of a chaperone have been suggested and appear likely to fulfill the role prokaryotes and are discussed below (Figure 1).
Hfq
Hfq was first identified as a Host factor for bacteriophage Qβ replication in E. coli (32,44). Hfq has been most closely associated with sRNA in Gram negative bacteria where it stabilized sRNAs and promotes their interactions with mRNA. The ternary complex of sRNA-mRNA-Hfq interacts with RNase, which is required for the cleavage of the sRNA-mRNA duplex facilitated by Hfq (49,50). Deletion of Hfq has been associated with sRNA mediated activity such as diminished stress tolerance, growth defects, and impaired virulence in pathogens (51-54).
Hfq is a member of the Sm-like family of proteins LSm (55) consisting of a homohexamer, with each monomer consisting of an LSm-domain (50,56,57). The homohexamers of Hfq form a ring structure ~65 Å in diameter (57). The ring structure of Hfq consists of two distinct faces indicated as the proximal and distal faces, and the outside ring surface referred to as the rim or lateral surface (32,50). The proximal face of Hfq has sites specific binding sites for polyU sequences, which is conserved in Gram-negative and Gram-positive bacteria. The proximal face of Hfq binds to the polyU tail of the Rho-independent terminator sequence of sRNAs and acidic residues on the proximal face may help discriminate between sRNA and other cellular RNAs (32).
The distal face of Hfq binds RNA sequences that are A-rich with a species specific preference. Gram-negative bacteria prefer a poly-(ARN) sequence and Gram-positive bacteria prefer a poly-(AN) sequence, where R is a purine and N is any nucleotide (32,50). The binding site on the distal face of Hfq binds to the poly-A tail of the mRNA. The mRNA will only bind to the Hfq if the complementary region with the sRNA does not overlap with the Hfq binding sequence of the mRNA. Additionally, the complementary sequence of the mRNA must be properly spaced from the Hfq binding sequence. The secondary structure of the mRNA in some instances aids in proper positioning of the Hfq binding sequence and the sRNA complementary sequence of the mRNA to allow for proper alignment and interactions with each (32).
The rim of Hfq contains positively charged residues on the outside of grooves on each of the six monomers of Hfq. These positively charged residues act as a secondary binding site for AU rich sequences of sRNA via the phosphate backbone, and the distance between the AU rich sequences of the sRNA and the complementary sequence of the sRNA and mRNA effects regulation (58). The rim of Hfq has been proposed to facilitate the ‘seeding’ of the interactions between the strands of the sRNA and the mRNA target (48). The seed sequence of the sRNA is hypothesized to be presented as a single strand by Hfq and the sRNA/Hfq complex then probes the mRNA in search of a compliment to the seed sequence.
Hfq impacts multiple steps in facilitating the interactions of mRNA and sRNA by changing the structures of RNA, bringing RNAs into proximity of each other to interact, neutralizing negative charge, stimulating nucleation of the first base pair, as well as further annealing of base pairs (32). What is less defined is how Hfq finds the correct sRNAs to be bound. A study of the diffusion rate of Hfq has shown that Hfq binds at least some RNA when it is being transcribed, but also binds RNAs at times other than during transcription (59). Additionally, the optimal sRNA regulation is Hfq concentration dependent, sRNA concentration dependent, mRNA concentration dependent, and depends also on the concentration of “decoy” sRNA targets such as fragments of processed tRNAs and bacteriophage transcripts (44).
Argonaute
Ago proteins are present in all kingdoms of life and are known to be a key factor in RNA silencing in eukaryotes. Crystal structures of eukaryotic human Ago (eAgo), and prokaryotic Pyrococcus furiosus and Aquifex aeolicus (pAgos) show conserved structures in prokaryotic and eukaryotic Ago proteins (60-64). All structures contain N-terminal is a PIWI-Argonaute-Zwille (PAZ) domains in one lobe of the bi-lobed structure, while middle (MID) and the RNas H-like PIWI domains form the other lobe. Both pAgos and eAgos bind ds nucleic acid structures, however, the shapes of the binding channel between pAgos and eAgos are not conserved. As a result, the pAgos have a preferential binding to B-form shaped structures of nucleic acid over A-form shaped structures. The deviations in the shapes of the binding channels between the different Ago proteins explains the preferential association of pAgo with formation of dsDNA structures and dsDNA-RNA hybrid structures as opposed to eAgo proteins which preferentially chaperone the formation of dsRNA structures (62,64). Therefore, the role of Ago proteins may have evolved to form a RISC-type complex in eukaryotes. However, while the preference for Ago proteins association with RNAs into the formation of a RISC-like structure in prokaryotes seems unlikely based on the current data, future research may identify a pAgo that can chaperon the formation of an RNA duplex in prokaryotes.
YbeY
YbeY has a conserved MID domain similar to Ago proteins, and has been proposed to act as chaperone in sRNA regulation (49,65). Hfq has not been identified in all sequenced bacteria genomes. However, YbeY has been found to be one of the genes that comprise the minimal bacterial genome. YbeY has been shown to influence maturation of rRNA and be involved in the quality control of the 70s ribosome (49). In addition to having a MID domain similar to Ago proteins, the crystal structure of YbeY contains a metal ion coordinated by three histidines and has been shown to possess single-strand nuclease activity (66). Additionally, mutant strains of YbeY have similar phenotypes as Hfq mutant strains (65). YbeY has been shown to regulated sRNAs in E. coli in response to hydroxyurea and modulate Hfq-dependent and Hfq-independent sRNAs. These results have led to the proposal that YbeY acts in concert with Hfq in bacteria that is highly depedent on sRNA regulation and independent of Hfq in bacteria strains that lack Hfq for sRNA regulation (49).
Non-miRNA like regulation by asRNAs
The mechanism of regulation of mRNA translation by asRNAs is similar to that of sRNAs in that the initial interactions occur in the seed regions of the asRNA and the mRNA (6). The seed sequence interaction often occurs by loop-loop kissing interactions between two loops of the asRNA and two loops of the mRNA, but interactions between single loops and a loop and a single strand have also been reported with asRNA. The seed regions of asRNAs are typically CG rich (6), and an U-turn motif, similar to that of the anticodon sequence of tRNAs, is conserved in the loop regions of many asRNAs and in the interacting loop regions of the target mRNA (67,68). The U-turn contains a conserved YUNR (Y = pyrimidine, N = any nucleotide, R = purine) in the loop region has been associated with asRNA seed sequence (67).
Unlike eukaryotic miRNAs and prokaryotic sRNAs, the binding of asRNAs to their mRNA targets normally occurs without the aid of chaperone proteins (3,6,8). The independence of asRNAs from chaperone proteins likely arises from the location that they originate on the antisense strand of their target mRNA. The origins of asRNA dictate that they are perfectly complimentary to the mRNA. Additionally, asRNAs typically have greater overlap of complementary sequence with the target mRNA. These two factors combined would result in a more stable and more persistent asRNA/mRNA complex compared to a complex between the sRNA and mRNA.
A more important factor in the asRNA chaperone independent binding of the mRNA target may be its proximity to its mRNA target upon transcription. Since, both the mRNA and asRNA are transcribed from the same loci, the local concentration of the asRNA relative to the mRNA target is higher. The importance of the initial localization of the asRNA with its target mRNA is demonstrated by the silencing of genes in E. coli by short perfectly complimentary sequences introduced by an expression plasmid that are trans-encoded to the target mRNA (10). In this work, a 400 nt transcript was required to silence the mRNA target. Most naturally occurring asRNAs require significantly fewer nt complements to bind the target mRNA. While it should be noted that the sequences used were not optimized for seed sequence containing secondary structures that may lead to increased efficiency in mRNA silencing for shorter sequences, the work demonstrates that the complementarity between mRNA and asRNAs is not the primary factor in their ability to silence mRNA independent of chaperone proteins. Additionally, the importance of co-localization of sRNAs with the mRNA targets in an Hfq-dependent mRNA silencing has been demonstrated with membrane associated mRNA, ptsG, and the co-localized sRNA, SgrS (69). It has been previously noted that the localization patterns of mRNA and how it relates to silencing by sreRNAs may play an important factor and should garner more attention (6).
Applications of sreRNAs
The investigation of regulating RNAs extends our understanding of new cellular pathways in bacteria and opens the potential for new genomic tools. Additionally, regulatory RNAs have recently been associated with antibiotic resistance in pathogenic bacteria. Pseudomonas putid strain DOT-T1E has demonstrated a wide range of antimicrobial resistance. Exposure of this strain to a wide spectrum of antibiotics leads to the production of 138 novel sRNAs by the bacteria. While exact mechanism of the sRNAs produced by the bacteria was not identified, the production of these sRNAs was coupled with the up and down regulation of the genes that were specific to the stress caused by a specific antibiotic (70).
The resistance of Staphylococcus aureus strains to antimicrobial glycopeptides have been shown to be an associated with cell wall thickening in response to the presence of these antibiotics (71). A DNA binding protein, SpoVG, has been shown to regulate the operon associated with this response. The processing of the mRNA that codes for the SpoVG is silenced by sRNA, SprX. The silencing occurs by the recognition of SprX encoded UCCC region located in an exposed loop region of the sRNA and a single strand sequence in the ribosomal binding domain of SpoVG mRNA in a Hfq independent mechanism. The binding of SprX inhibits the translation of SpoVG mRNA. The silencing of the SpoVG mRNA results in an increase in antibiotic resistance (71). Therefore, SprX is expressed as a response to glycopeptide antibiotics and could be a potential therapeutic target to increase the effect of these antibiotics in resistant S. aureus strains.
In addition to the previously mentioned links between sRNAs and antibiotic resistance, other regulating RNAs have been determined or suggested to influence antibiotic resistance. Particularly, regulatory RNAs associated with RNA synthesis, protein synthesis, cell membrane integrity, cell wall turnover, and the regulation of membrane proteins have all been associated with antibiotic resistance in a variety of bacteria (72,73). The association of regulatory RNAs with a wide variety of cellular pathway, and in particularly responses to stress and antibiotic resistance may provide a new target for therapeutic agents. The use of RNA interference (RNAi) technology may be one strategy in the development of these new agents. However, sRNAs are not highly conserved and therefore, would target only a small number of target pathogens (74). Alternative targets may involve the proteins associated with regulatory RNAs. For instance, the gene coding for Hfq is present in over 50% of bacteria genomes sequenced. A library of cyclic peptides was used to find a compound that inhibited sRNA binding by Hfq in vitro. The same peptide was shown to inhibit the regulation of two different sRNAs that associate with Hfq in vivo (36). Further understanding of the sRNA pathways has the potential for additional discovery and development of therapeutic agents that act as antibiotics or reduce the resistance of pathogenic bacteria to existing antibiotics.
Conclusions
Similar to the discovery of the role of CRISPR and miRNAs in RNA interference, the role of sreRNAs may give rise to new genomic tools, a better understanding of cellular pathways and new therapeutic agents. The use of Next Generation RNA sequencing has already allowed the identification of a large number of sreRNAs and potential sreRNAs, and research is underway to identify more (7). One of the immediate challenges is classification of sreRNA into groups that work by similar mechanisms. One such classification presented here is to group the sreRNAs based on the definition of asRNA and sRNA. This classification is based on the difference in complementarity and the requirement of chaperone proteins that typically accompanies these RNAs. However, reports of asRNAs that are associated with Hfq (37), and sRNAs, such as SprX, that silence mRNA independent of Hfq (71), indicate that this classification will likely overlap. As future research identifies common structural elements, associated proteins, common origins and processing, and mechanisms of action, similar to the well-defined characteristics of miRNAs, the categories of different sreRNAs should become more apparent. This categorization should lead to a more refined approach to the investigation of regulatory RNAs.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.10.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Saberi F, Kamali M, Najafi A, et al. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. 2016; Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5415839/. Accessed 7/23/2019, 2019.
- Lybecker M, Bilusic I, Raghavan R. Pervasive transcription: detecting functional RNAs in bacteria. 2014; Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4581347/. Accessed 7/23/2019, 2019.
- Harris KA, Breaker RR. Large Noncoding RNAs in Bacteria. Microbiol Spectr 2018; [Crossref] [PubMed]
- Khorkova O, Myers AJ, Hsiao J, et al. Natural antisense transcripts. 2014; Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4170719/. Accessed 7/23/2019, 2019.
- Lioliou E, Romilly C, Romby P, et al. RNA-mediated regulation in bacteria: from natural to artificial systems. N Biotechnol 2010;27:222-35. [Crossref] [PubMed]
- Lejars M, Kobayashi A, Hajnsdorf E. Physiological roles of antisense RNAs in prokaryotes. Biochimie 2019;164:3-16. [Crossref] [PubMed]
- Lasa I, Toledo-Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. 2012; Available online: https://www-ncbi-nlm-nih-gov.ezproxy3.lhl.uab.edu/pmc/articles/PMC3551857/. Accessed 7/23/2019, 2019.
- Rosikiewicz W, Makalowska I. Biological functions of natural antisense transcripts. Acta Biochim Pol 2016;63:665-73. [PubMed]
- Magistro G, Magistro C, Stief CG, et al. A simple and highly efficient method for gene silencing in Escherichia coli. J Microbiol Methods 2018;154:25-32. [Crossref] [PubMed]
- Lee YJ, Kim SJ, Amrofell MB, et al. Establishing a Multivariate Model for Predictable Antisense RNA-Mediated Repression. ACS Synth Biol 2019;8:45-56. [Crossref] [PubMed]
- Inouye M. Antisense RNA: its functions and applications in gene regulation--a review. Gene 1988;72:25-34. [Crossref] [PubMed]
- Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol 2014;11:1335-9. [Crossref] [PubMed]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 2004;14:647-56. [Crossref] [PubMed]
- Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet 2005;21:260-4. [Crossref] [PubMed]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell 2009;136:615-28. [Crossref] [PubMed]
- Stazic D, Pekarski I, Kopf M. A Novel Strategy for Exploitation of Host RNase E Activity by a Marine Cyanophage. Genetics 2016;203:1149-59. [Crossref] [PubMed]
- Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 2011;39:4890-9. [Crossref] [PubMed]
- Melior H, Li S, Madhugiri R, et al. Transcription attenuation-derived small RNA rnTrpL regulates tryptophan biosynthesis gene expression in trans. Nucleic Acids Res 2019;47:6396-410. [Crossref] [PubMed]
- Kolb FA, Engdahl HM, Slagter-Jager JG, et al. Progression of a loop–loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. 2000; Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC305787/. Accessed 7/25/2019, 2019.
- Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 2007;21:1353-66. [Crossref] [PubMed]
- Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J 1990;9:1391-9. [Crossref] [PubMed]
- Huntzinger E, Boisset S, Saveanu C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 2005;24:824-35. [Crossref] [PubMed]
- Morfeldt E, Taylor D, von Gabain A, et al. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 1995;14:4569-77. [Crossref] [PubMed]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 2004;186:6698-705. [Crossref] [PubMed]
- Brunel C, Marquet R, Romby P, et al. RNA loop-loop interactions as dynamic functional motifs. Biochimie 2002;84:925-44. [Crossref] [PubMed]
- Franch T, Petersen M, Wagner EG, et al. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol 1999;294:1115-25. [Crossref] [PubMed]
- Thisted T, Sorensen NS, Wagner EG, et al. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5'-end single-stranded leader and competes with the 3'-end of Hok mRNA for binding to the mok translational initiation region. EMBO J 1994;13:1960-8. [Crossref] [PubMed]
- Mars RA, Mendonca K, Denham EL, et al. The reduction in small ribosomal subunit abundance in ethanol-stressed cells of Bacillus subtilis is mediated by a SigB-dependent antisense RNA. Biochim Biophys Acta 2015;1853:2553-9. [Crossref] [PubMed]
- André G, Even S, Putzer H, et al. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 2008;36:5955-69. [Crossref] [PubMed]
- Chen J, Morita T, Gottesman S. Regulation of Transcription Termination of Small RNAs and by Small RNAs: Molecular Mechanisms and Biological Functions. Front Cell Infect Microbiol 2019;9:201. [Crossref] [PubMed]
- Updegrove TB, Zhang A, Storz G. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 2016;30:133-8. [Crossref] [PubMed]
- Lasa I, Toledo-Arana A, Dobin A, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 2011;108:20172-7. [Crossref] [PubMed]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys 2013;42:217-39. [Crossref] [PubMed]
- Rusk N. Prokaryotic RNAi. Nat Methods 2012;9:220-1. [Crossref] [PubMed]
- El-Mowafi SA, Alumasa JN, Ades SE, et al. Cell-based assay to identify inhibitors of the Hfq-sRNA regulatory pathway. Antimicrob Agents Chemother 2014;58:5500-9. [Crossref] [PubMed]
- Bilusic I, Popitsch N, Rescheneder P, et al. Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol 2014;11:641-54. [Crossref] [PubMed]
- Kang SM, Choi JW, Lee Y, et al. Identification of microRNA-size, small RNAs in Escherichia coli. Curr Microbiol 2013;67:609-13. [Crossref] [PubMed]
- Lee HJ, Hong SH. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol Lett 2012;326:131-6. [Crossref] [PubMed]
- Furuse Y, Finethy R, Saka HA, et al. Search for microRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS One 2014;9:e106434 [Crossref] [PubMed]
- Bloch S, Wegrzyn A, Wegrzyn G, et al. Small and Smaller-sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins (Basel) 2017; [Crossref] [PubMed]
- Nicholson RH, Nicholson AW. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm Genome 2002;13:67-73. [Crossref] [PubMed]
- Ma E, Zhou K, Kidwell MA, et al. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol 2012;422:466-76. [Crossref] [PubMed]
- Papenfort K, Said N, Welsink T, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 2009;74:139-58. [Crossref] [PubMed]
- Guo MS, Updegrove TB, Gogol EB, et al. MicL, a new sE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. - PubMed - NCBI. 2014; Available online: https://www.ncbi.nlm.nih.gov/pubmed/?term=micl+new+dependent+srna+stress+lpp+lipoprotein. Accessed 7/24/2019, 2019.
- MacRae IJ, Ma E, Zhou M, et al. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A 2008;105:512-7. [Crossref] [PubMed]
- Rand TA, Ginalski K, Grishin NV, et al. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci U S A 2004;101:14385-9. [Crossref] [PubMed]
- Gorski SA, Vogel J, Doudna JA. RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol 2017;18:215-28. [Crossref] [PubMed]
- Pandey SP, Winkler JA, Li H, et al. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics 2014;15:121. [Crossref] [PubMed]
- Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol 2013;10:610-8. [Crossref] [PubMed]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 1994;13:35-49. [Crossref] [PubMed]
- Pannekoek Y, Huis in 't Veld R, Hopman CT, et al. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol Lett 2009;294:216-24. [Crossref] [PubMed]
- Sittka A, Pfeiffer V, Tedin K, et al. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 2007;63:193-217. [Crossref] [PubMed]
- Geng J, Song Y, Yang L, et al. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS One 2009;4:e6213 [Crossref] [PubMed]
- Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol 2004;344:1211-23. [Crossref] [PubMed]
- Horstmann N, Orans J, Valentin-Hansen P, et al. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res 2012;40:11023-35. [Crossref] [PubMed]
- Schumacher MA, Piro KM, Xu W, et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009;323:396-401. [Crossref] [PubMed]
- Ishikawa H, Otaka H, Maki K, et al. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3' poly(U) tail. RNA 2012;18:1062-74. [Crossref] [PubMed]
- Persson F, Lindén M, Unoson C, et al. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat Methods 2013;10:265. [Crossref] [PubMed]
- Yuan YR, Pei Y, Ma JB, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 2005;19:405-19. [Crossref] [PubMed]
- Rivas FV, Tolia NH, Song JJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 2005;12:340-9. [Crossref] [PubMed]
- Doxzen KW, Doudna JA. DNA recognition by an RNA-guided bacterial Argonaute. PLoS One 2017;12:e0177097 [Crossref] [PubMed]
- Ender C, Meister G. Argonaute proteins at a glance. J Cell Sci 2010;123:1819-23. [Crossref] [PubMed]
- Ryazansky S, Kulbachinskiy A, Aravin AA. The Expanded Universe of Prokaryotic Argonaute Proteins. MBio 2018; [Crossref] [PubMed]
- Pandey SP, Minesinger BK, Kumar J, et al. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res 2011;39:4691-708. [Crossref] [PubMed]
- Zhan C, Fedorov EV, Shi W, et al. The ybeY protein from Escherichia coli is a metalloprotein. Acta Crystallogr Sect F Struct Biol Cryst Commun 2005;61:959-63. [Crossref] [PubMed]
- Franch T, Gerdes K. U-turns and regulatory RNAs. Curr Opin Microbiol 2000;3:159-64. [Crossref] [PubMed]
- Asano K, Niimi T, Yokoyama S, et al. Structural basis for binding of the plasmid ColIb-P9 antisense Inc RNA to its target RNA with the 5'-rUUGGCG-3' motif in the loop sequence. J Biol Chem 1998;273:11826-38. [Crossref] [PubMed]
- Kawamoto H, Morita T, Shimizu A, et al. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev 2005;19:328-38. [Crossref] [PubMed]
- Molina-Santiago C, Daddaoua A, Gomez-Lozano M, et al. Differential transcriptional response to antibiotics by Pseudomonas putida DOT-T1E. - PubMed - NCBI. 2015; Available online: https://www.ncbi.nlm.nih.gov/pubmed/25581266. Accessed 7/24/2019, 2019.
- Eyraud A, Tattevin P, Chabelskaya S, et al. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res 2014;42:4892-905. [Crossref] [PubMed]
- Lalaouna D, Eyraud A, Chabelskaya S, et al. Regulatory RNAs involved in bacterial antibiotic resistance. PLoS Pathog 2014;10:e1004299 [Crossref] [PubMed]
- Dar D, Sorek R. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Curr Opin Microbiol 2017;36:111-7. [Crossref] [PubMed]
- Colameco S, Elliot MA. Non-coding RNAs as antibiotic targets. Biochem Pharmacol 2017;133:29-42. [Crossref] [PubMed]
Cite this article as: Watkins D, Arya DP. Regulatory roles of small RNAs in prokaryotes: parallels and contrast with eukaryotic miRNA. Non-coding RNA Investig 2019;3:28.


