
Long non-coding RNAs in placental development and disease
Introduction
The central dogma of conventional biological science revolves around the production of futuristic proteins as solitary trans-actors of genomic information. However, completion of human genome sequencing as well as high-throughput sequencing of transcriptome revealed that nearly 98% of the genome generates non-coding transcripts. Thousands of such non-coding RNAs have been reported in drosophila (1,2), C. elegans (3), zebrafish (4), mouse (5,6) and humans (7,8). Of these, long non-coding RNAs (lncRNA) contribute to the largest proportion of the mammalian non-coding transcriptome and play diverse regulatory roles that include gene expression (9,10), differentiation (11-13), development (14,15) as well as pathogenesis of several diseases (8,16) including various types of cancers (17).
In Eutherian mammals, proper fetal growth and development within mother’s body is dependent on the ability of the fetus to maintain a balance between the adequate supply of nutrients and simultaneous disposal of waste materials to the maternal circulation. The organ facilitating this bi-directional exchange process is the placenta (18,19). Placental morphogenesis involves differentiation of the multi-potent trophoblast stem cells into various trophoblast lineages as well as extensive angiogenesis (20-25). Trophoblast cells assume various specialized functions of the placenta throughout gestation. Inadequate development of trophoblast cells results in adverse pregnancy outcome affecting both the fetus and the mother. These include various placenta associated disorders, such as, intra-uterine growth restriction, pre-eclampsia, whose etiologies are not sufficiently understood (26-29).
Placenta harbors a unique epigenome (30-33) exclusive from other somatic tissues of the body. It is therefore likely that long non-coding RNAs would play significant role in trophoblast development leading to a functional placenta. This review is an integrated summary on the prevailing lncRNA research central to the development of the placenta optimized to provide their significance to perinatal medicine.
Long non-coding RNAs
Characteristics of lncRNAs
LncRNAs share many features of protein-coding RNAs, including 5’-capping, splicing, poly-adenylation as well as epigenetic marks in the promoter such as increased H3K4me3 and RNA Pol-II binding site. Interestingly, some lncRNAs possess H3K4me1 marks indicating transcription from active enhancer sites (34). Non-polyadenylated lncRNAs are found predominantly in the nucleus. Absence of lncRNA homolog across species indicates lack of sequence conservation. However, sequence conservation in promoter regions of lncRNAs is similar to protein-coding transcripts (34,35). Folding of lncRNAs are rather complex and they form secondary and tertiary structures to serve their functions (36). Owing to their vast numbers, many of newly discovered lncRNAs are poorly characterized with respect to their functions.
Classification of lncRNAs
There are many reviews (37-39) on classification of lncRNAs as classification eases hypothesis-driven research on function of newly discovered lncRNAs. LncRNAs are primarily classified based on (I) genomic locations, (II) functions.
Depending on the genomic locations with respect to the protein-coding genes, lncRNAs can be categorized into the following biotypes:
- Long intergenic non-coding RNA (lincRNA): This group of lncRNAs does not intersect with the coding region of any gene. Indeed, they are encoded in between reported protein-coding genes.
- Divergent lncRNA, also called pancRNA (promoter associated non-coding RNA): They are transcribed from the opposite strand of the same promoter region of protein coding gene.
- Convergent lncRNA: They are transcribed from opposite strands in reverse direction facing each other.
- Intronic lncRNA: They are transcribed from introns.
- Overlapping sense lncRNA: They are encoded from sequence overlapping with the sense strand of other gene.
- Overlapping anti-sense lncRNA: They are encoded from sequence overlapping with the anti-sense strand of other gene.
- Enhancer lncRNA: They are transcribed from enhancer regions of other genes and may be expressed as either uni or bi-directional transcripts.
Functions of lncRNAs
Experimentally determined functions of lncRNAs include modification of the chromatin, regulation of transcription and RNA processing, regulation of imprinting and formation of ribo-nucleoprotein complex. There are many elegant reviews that detail functional classification of lncRNAs (40-42). We will briefly describe the functional classification here. Depending on the molecular function lncRNAs can be categorized into the following archetypes (Figure 1):
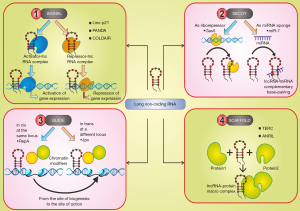
- As signals: Due to their cell type-specific as well as unique spatiotemporal expression pattern, lncRNAs may integrate developmental cues and also respond to various biological stimuli and thus function as molecular signals in a variety of developmentally regulated processes. Examples of such lncRNAs include (I) linc-p21 (43) and PANDA (44), which are induced by DNA damage; (II) ROR lncRNA induced by Oct-4 depletion (45) in human iPS cells, (III) plant lncRNA, COLDAIR and COOLAIR are induced by cold and can silence the flowering locus C transcription and promotes Polycomb occupancy in Arabidopsis (46).
- As decoys: They act as “molecular sink” by binding to and titrating target proteins such as RNA-binding proteins (RBPs), transcription factors, or chromatin modifiers or a RNA, in particular miRNAs. Examples include (I) Gas5 lncRNA that impart glucocorticoid resistance (47); (II) Cyrano lncRNA act as a miR-7 sponge in embryonic stem cells controlling self-renewal (48).
- As guide: They bind to and direct ribonucleoprotein complexes either in cis- or trans- to specific target locations to regulate gene expression. One example of lncRNA acting as cis-guide is lncRNA RepA-mediated PRC2 recruitment and H3K27 trimethylation of the Xist promoter leading to transcriptional induction of Xist (49). One example of lncRNA acting as trans-guide include Jpx lncRNA that trans-activates Xist although the precise mechanism remains elusive (50).
- As scaffolds: They serve as molecular podium on which protein or RNA moieties can be assembled at the same type and space into a macromolecular complex. One example of scaffold is lncRNA is TERC, which is known to assemble telomerase (51).
It is imperative that a single lncRNA might possess multiple functions as described above based on availability as well as cellular context.
Discovery of lncRNAs
Low abundance, tissue specificity and lack of sequence conservation make lncRNA discovery difficult. Yet, there are a number of cutting edge methodologies that are being used leading to discovery of lncRNAs. These include (I) cDNA library and tiling arrays, (II) high throughput sequencing serial analysis of gene expression (SAGE), (III) RNA sequencing (RNA-seq), (IV) Cap analysis of gene expression (CAGE), (V) Global run-on sequencing (Gro-seq), (VI) 5'-bromo-uridine immunoprecipitation chase-deep sequencing analysis (bric-seq).
cDNA library and tiling arrays
Identification of lncRNAs were done initially by generation and sequencing of cDNA libraries. The Functional Annotation of the Mammalian Genome (FANTOM) consortium led by RIKEN Omics Science Center has provided the largest collection of about 60770 full-length cDNA sequences for several species (52-54) along with a large number of lncRNAs (52-55).
Another classical method to identify lncRNAs is using tiling arrays. In this method, microarray slides, carrying tiled oligonucleotides that can be designed to cover non-repetitive sequences of specific chromosomes or, potentially, the entire genome, are used to hybridize cDNAs. Tiling arrays used to hybridize polyadenylated transcripts from primary human fibroblasts led to discovery of many lncRNAs including HOTAIR. Emergence of next generation sequencing (NGS) technologies made tiling arrays obsolete due to various drawbacks of using microarrays.
High throughput sequencing serial analysis of gene expression (SAGE)
High-throughput analysis of transcriptome was first done through SAGE. It involves generation of SAGE tags (short cDNA sequence) containing restriction enzyme recognition site at the 3' end of transcripts. SAGE tags are concatenated, cloned and sequenced to identify transcripts. LncRNAs in human cancerous tissues were identified across all 22 autosomes and the sex chromosomes using this method (56). SAGE has been replaced by NGS technologies currently as it profiles a larger number of transcripts in greater depth.
RNA sequencing (RNA-seq)
RNA-seq is the most widespread method used for novel lncRNA discovery. Construction of cDNA sequencing library by using random primers on rRNA-depleted transcripts allows sequencing of both poly(A) + and poly(A) – transcripts. Huge number and variety of lncRNAs in mouse ES cells, neuronal precursor cells, lung fibroblasts, and human tissue/cells have been discovered in using RNAseq (57-59). RNA capture sequencing that combines tiling arrays with RNA-seq led to discovery of many lncRNAs in fibroblasts (60).
Cap analysis of gene expression (CAGE)
CAGE allows simultaneous mapping and quantification of 5'-capped RNAs and is an effective tool for the identification of transcriptionally active promoter regions and Pol II-driven transcription start sites (TSS).
Single cell transcriptional profiling can be done using nanoCAGE. Using CAGE, genome wide comparison of transcripts have been done on human and mouse induced pluripotent stem (iPS) cells, embryonic stem (ES) cells, and differentiated cells (61). About 8,000 novel transcripts were thus identified in mice and 4,000 in humans. These were unannotated lncRNAs that overlap with regulatory regions and associate with retrotransposon elements.
Global run-on sequencing (Gro-seq)
GRO-seq method is based on nuclear run-on assay in the presence of Br-UTP labeled nucleotides followed by capture of Br-UTPtagged transcripts with the antibody anti-Br-UTP and subsequently deep sequencing. This method sequences nascent RNAs and thus provides a genome-wide location, orientation, and density of Pol II-engaged transcripts at a high resolution. Using Gro-seq in human ES cells, antisense lncRNA that arise by divergent transcription were discovered (62). GRO-seq analyses have revealed that only a small fraction of lncRNAs produced by divergent transcription is stable as divergent lncRNA transcripts are generally subjected to exosome-mediated degradation (63).
5'-bromo-uridine immunoprecipitation chase-deep sequencing analysis (bric-seq)
In BRIC-seq endogenous transcripts are labeled by adding 5'-bromo-uridine (BrU) to cell culture media. From isolated total RNAs, BrURNAs are recovered using immuno-purification, which is followed by RT-qPCR or deep sequencing. Fifteen new lncRNAs has been discovered by BRIC-seq on HeLa cells (64). Three of these lncRNAs are involved in cell proliferation and have been named short-lived noncoding transcripts (SLiTs).
Web resource for lncRNA research
So far, many web-based resources for lncRNAs have been reported. Table 1 summarizes online resources of lncRNAs.
Table1
| Name of the database | Description | URL | Reference |
|---|---|---|---|
| lncrnadb | Reference database containing a list of potential functionally relevant lncRNAs | http://www.lncrnadb.org/ | (65) |
| NONCODE | It is an integrated database dedicated to non-coding RNAs especially lncRNAs (excluding rRNA and tRNAs) of 17 different species | http://www.noncode.org/ | (66) |
| lncRNASNP2 | Database for the mutations and single nucleotide polymorphisms affecting lncRNA structure and function | http://bioinfo.life.hust.edu.cn/lncRNASNP2 | (67) |
| Lnc2Meth | A manually curated database to uncovering the biological relationship between lncRNAs and DNA methylation in disease complications | http://bio-bigdata.hrbmu.edu.cn/Lnc2Meth/ | (68) |
| EVLncRNAs | The currently largest database of experimentally validated repertoire of lncRNAs from 77 different species | http://biophy.dzu.edu.cn/EVLncRNAs/ | (69) |
| DES-ncRNA | A knowledgebase containing data-mined information on human lncRNA research | http://www.cbrc.kaust.edu.sa/des_ncrna/home/index.php | (70) |
| lncATLAS | A comprehensive database of lncRNA localization based on RNA sequencing datasets | http://lncatlas.crg.eu/ | (71) |
| FARNA | A knowledgebase of inferred functions of non-coding RNA transcripts in humans | http://cbrc.kaust.edu.sa/farna | (72) |
| lncSNP 2.0 | A database of the single nucleotide polymorphisms in human lncRNAs | http://bioinfo.hrbmu.edu.cn/LincSNP | (73) |
| lncVar | A database associated with the genetic variations associated with lncRNAs in 6 different species | http://bioinfo.ibp.ac.cn/LncVar/ | (74) |
| NPInter v3.0 | A database of the experimentally verified interaction between lncRNAs and other molecules | https://www.bioinfo.org/NPInter/ | (75) |
| deepBase v2.0 | A database decoding the evolution, expression patterns and functions of lncRNAs across 19 species | http://rna.sysu.edu.cn/deepBase/ | (76) |
| lncReg | A comprehensive database highlighting the regulatory networks associated with lncRNAs | http://bioinformatics.ustc.edu.cn/lncreg/ | (77) |
| LNCipedia | A reference database for annotated human lncRNA sequences | https://lncipedia.org/ | (78) |
| LncRNA2Target | A comprehensive database of differentially expressed genes after lncRNA knockdown or over-expression | https://omictools.com/lncrna2target-tool | (79) |
| lncRNome | A knowledgebase of human lncRNAs | http://genome.igib.res.in/lncRNome/ | (80) |
| lnCeDB | Database of human lncRNAs functioning as competing endogenous RNAs | http://gyanxet-beta.com/lncedb/ | (81) |
| CPC | A machine-based classifier to predict the protein coding potential of input transcript sequences | http://cpc.cbi.pku.edu.cn/ | (82) |
| COME | A multi-featured database for identification and characterization of novel lncRNAs | https://github.com/lulab/COME | (83) |
| CPAT | An alignment-free logistic regression-based method which discriminates non-coding transcripts from coding transcripts | http://lilab.research.bcm.edu/cpat/index.php | (84) |
| PNRD | An integrated database for searching, browsing, predicting and visualizing plant non-coding RNA | http://structuralbiology.cau.edu.cn/PNRD | (85) |
Placenta
Development of placenta
Placenta is an extra-embryonic transient endocrine organ that develops during pregnancy in Eutherian mammals. It originates by differentiation and morphogenesis from a single layer of trophectoderm that lines the outer layer of blastocyst. Although mouse and human placenta have similarities in terms of function, there exist striking differences in trophoblast cell subtypes in the placenta (Figure 2).
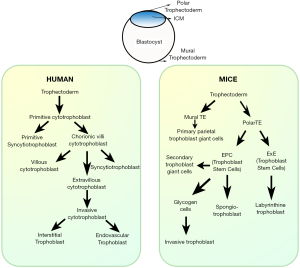
In humans, implantation occurs 7–8 days following fertilization. As implantation is initiated maternal uterine endometrial stromal cells (fibroblast-like) differentiate into the decidua by action of hormone progesterone as well as embryonic stimuli. The entire maternal decidua is divided into three regions: decidua basalis, decidua capsularis and decidua parietals (decidua vera). Implantation takes place at the decidua basalis and the basal plate is formed in this region. This can be subdivided into a zona compacta and a zona spongiosa (where the detachment of the placenta takes place following birth). Decidua capsularis lies like a capsule around the chorion. Decidua parietalis is located on the opposite uterus wall.
Following attachment the polar trophectoderm cells of the blastocyst differentiate into multi-nucleated syncytiotrophoblast cells that invade the decidua basalis. Lacunae, formed within the syntiotrophoblast layer, connect with each other and are filled by maternal blood from eroded decidual blood vessels. These events happen around 9–10th embryonic day. Proliferating cytotrophoblast cells of the chorion penetrate into the cords of the syncytiotrophoblast creating the primary trophoblast villi between the 11th and 13th day (86). After the 16th day the mesenchyme, developed from extra-embryonic mesoderm of the chorion fill the villi, which is now called a secondary villi. At the end of the 3rd week, the mesenchyme of the villi differentiates to form connective tissue and blood vessels. Villi that contain differentiated blood vessels are called tertiary villi. The villi morph into a treelike shape consisting of a mesenchymal core containing fetal circulation and are surrounded by a double trophoblast layer, the outer syncytiotrophoblast, and the inner cytotrophoblast. The intravillous vascular network connects with the umbilical-allantoic vessels establishing the fetal placental circulation. The intra-villous space contains the lacunae, which are already sites of intense maternal circulation. The villi that attaches with the maternal tissue are called anchoring villi. The others remain free in the intervillous spaces and are referred to as floating villi. Around 6 weeks of gestation, some cytotrophoblast cells of the anchoring villi leave the placenta and penetrate through the decidua deep into uterine myometrium. These trophoblast cells are called extra-villous cytotrophoblast or invasive trophoblast cells (87). These invasive trophoblast cells also penetrate into the maternal spiral arteries and in effect remodel the spiral arteries making them flaccid and distended. Invasion of trophoblast cells into the decidua basalis, generate several wedge-shaped areas of decidua tissued called placental septa, which appear in month 4 starting out from the maternal (decidua) plate but do not reach the chorionic plate. These septa divide the fetal part of the placenta into 10–38 convex areas composed of lobes called cotyledons. Each cotyledon is made up of 2 or more anchoring villi and their many branches. Part of decidua basalis that is strongly adhered to the fetal placenta (Chorionic villi) is called maternal placenta and is ~0.5 cm thick. During pregnancy, the maternal blood volume increases by about 50% and the uterine blood flow increases 10 to 12-fold. Increase in flow happens due to spiral artery remodeling by trophoblast cells. Inadequate spiral artery remodeling leads to various placenta associated disorders.
In mice, implantation of the blastocyst is initiated by attachment of the mural trophectoderm to uterine epithelial cells. Following implantation trophectoderm cells differentiate and give rise to primary trophoblast giant cells (TGC) that encapsulate the entire conceptus except for the polar trophectoderm (88). The polar trophectoderm differentiates into ectoplacental cone and the extraembryonic ectoderm that form a cylindrical structure. Trophoblast stem cells (TSC) within the ectoplacental cone differentiate into the distinct regions of the junctional zone. TSCs located at the periphery of the ectoplacental cone differentiate into secondary TGCs that line the boundary of the decidua and placenta. TSCs also differentiate into spongiotrophoblast cells that form a sandwiched layer between the secondary trophoblast giant cells and the labyrinth zone. Glycogen cells of the junctional zone appear during the last half of pregnancy and are said to be the source of invasive trophoblast cells. Following the chorio-allantoic fusion, TSCs located within the extra-embryonic ectoderm differentiate to labyrinthine trophoblast cells and give rise to the labyrinth zone of the placenta. Two placental zone are evident in mice: junctional zone and labyrinth zone. Labyrinth zone may be compared to fetal placenta in human. Maternal decidua located right above the junctional zone slowly regresses as gestation progresses and part of maternal decidua remains strongly adhered to the placenta till the end of gestation.
Like in humans, a specialized population of trophoblast cells exit the placenta and invade the decidua and arteries. In mice trophoblast invasion is shallow and is limited to decidua, whereas, in rats and hamsters, invasion occurs deep into myometrium resembling that in humans. On time as well as adequate trophoblast invasion is absolutely necessary for healthy progression of pregnancy.
Placenta associated disorders
Pregnancy associated disorders most often arise due to insufficient placental development. Most prevalent of such disorders has been discussed below.
Intra-uterine growth restriction (IUGR)
IUGR is a pregnancy associated complication where the fetus is retarded from attaining its full gender and race specific genetic growth potential in utero. World Health Organization has defined low birth weight as one whose birth weight is less than 2,500 gms irrespective of gestational age. IUGR is said to be present in those babies whose birth weight is below tenth percentile of the average for the gestational age and it affects nearly 3–8% of worldwide pregnancies (89). Normal fetal growth is characterized by cellular hyperplasia followed by hyperplasia and hypertrophy and lastly by hypertrophy alone. Two third of fetal weight gain occurs beyond 24th week of pregnancy.
Types of IUGR
Pathological growth restricted fetuses can be of two types, (I) symmetrical (~30%) and (II) asymmetrical (~80%). Symmetric IUGR fetuses are affected from very early in the phase of cellular hyperplasia leading to less number of total cells. This form of IUGR is often caused by chromosomal abnormalities or congenital infection. Onset of asymmetrical IUGR generally occurs during phase of cellular hypertrophy. This type of IUGR generally develops due to maternal or placental causes extrinsic to the fetus.
Etiology of IUGR
It could be maternal, fetal placental or unknown. Maternal causes include malnutrition before and during pregnancy, maternal diseases, such as, anemia, hypertension, thrombophilia, heart disease, chronic renal disease, and substance abuse, such as, alcohol, smoking, drugs etc. Fetal causes of IUGR may include (I) chromosomal abnormality, which is associated with 8–12% of IUGR infants; (II) infection etc. Placental cause of IUGR most often could be devastating. It is caused by poor uterine blood flow for long time leading to inadequate substrate transfer. Primary cause of compromised placental blood flow has been attributed to inadequate spiral artery remodeling. Unknown causes of IUGR attribute to ~30% cases.
Complications
In the mother, fetal growth restriction per se does not cause any harm except when there is underlying causes like pre-eclampsia, heart disease, malnutrition, which may be life threatening. In the fetus, complications could be (I) antenatal that include chronic fetal distress, fetal death; (II) intranatal that include hypoxia and acidosis and (III) postnatal. Immediate postnatal complications may include asphyxia, hypoglycemia due to shortage of glycogen reserve in the liver, hypothermia, pulmonary hemorrhage, polycythemia, necrotizing enterocolitis due to reduced intestinal blood flow, Intraventricular hemorrhage. Late postnatal complications may include slow growth, which is found primarily in symmetrical growth retarded babies. It has been observed that IUGR babies are predisposed to develop cardiovascular disease, type2 diabetes and hyperlipidemia in adulthood.
Pre-eclampsia (PE)
PE is a multisystem disorder characterized by development of maternal hypertension and proteinuria after 20th week of gestation in a previously normotensive and non-proteinuric mother. Incidence of PE in varies widely from 2–10% worldwide. Pathophysiology of PE encompasses several organs/tissue that include (I) uteroplacental bed, (II) kidney, (III) blood vessels, (IV) liver.
Uteroplacental bed
There is increased evidence of premature ageing of the placenta with red/white infracts visible on the maternal surface of the placenta. There is syncytial degeneration, marked proliferation of cytotrophoblasts, thickening of basement layer, most importantly; normal endovascular trophoblast invasion is severely impaired, trophoblast invasion is shallow and is limited to decidua-myometrial junction. This results in maternal oxidative and endoplasmic reticulum stress. The blood flow to the placenta is decreased remarkably.
Kidney
Glomerular endotheliosis with swelling of endothelial cells and fibrin like deposits occur in the basement membrane. Interstitial cells in between the capillaries proliferate with associated spasm of the afferent glomerular arterioles. The net effects are reduced renal blood flow, glomerular filtration rate and impaired tubular resorption or secretory functions. Recovery is likely to be complete following delivery.
Blood vessels: There is intense vasospasm. Circulation in the vasa vasorum is impaired leading to damage in vascular walls as well as endothelial integrity.
Liver: Periportal hemorrhagic necrosis of the liver occurs due to thrombosis of the arterioles. However, hepatic insufficiency does not occur because of reserve capacity and regenerative ability of the liver cells.
Mitigation of the disease complications following the delivery of the placenta implicates PE as a placenta-associated pregnancy disorder.
Placenta accreta
In this rare disease placenta is directly anchored to the myometrium without any intervening decidua. The probable cause is defective decidual transformation. The condition is usually associated when the placenta is implanted in lower segment or over the previously injured sites as in Caesarean section. Pathological confirmation includes (I) absence of decidua basalis, (II) absence of Nitabuch’s fibrinoid layer and (III) carrying degree of penetration of the villi into the muscle bundle (increta) or upto serosal layer (percreta). Approximately 5–10% of post-partum hemorrhages are caused by placental accreta.
Gestational diabetes mellitus (GDM)
It is defined as abnormal carbohydrate tolerance with onset of or first detected during pregnancy. During normal pregnancy, early in gestation, maternal estrogen and progesterone increase and promote pancreatic β-cell hyperplasia and increased insulin release. In general, glucose tolerance deteriorates during pregnancy, but only 2–3% develops GDM. As pregnancy progresses, increased levels of human chorionic somatomammotropin (hCS), cortisol, prolactin, progesterone, and estrogen lead to insulin resistance in peripheral tissues, primarily in skeletal muscle. The mechanism of insulin resistance is likely a post-receptor defect, since normal insulin binding by insulin-sensitive cells has been demonstrated. The pancreas releases 1.5–2.5 times more insulin in order to respond to the resultant increase in insulin resistance. Mothers with normal pancreatic function are able to meet these demands, whereas, mothers with borderline pancreatic function have difficulty increasing insulin secretion and consequently produce inadequate levels of insulin leading to GDM. Therefore, GDM manifests as delayed or insufficient insulin secretion in the presence of increasing peripheral resistance in mothers. Children born to GDM mothers are at increased risk of metabolic syndrome. Women with GDM is disposed to increased risk of developing gestational hypertension and preeclampsia with long terms effects on developing metabolic syndrome (90) and cardiovascular disorders (91). Methods for diagnosis of GDM include fasting and screening of random plasma glucose in early pregnancy (92), oral glucose tolerance test (93) or using other serum markers like Pregnancy associated Plasma Protein-A (94), Placenta Growth factor (94) and endoglin (95). Therefore, future studies involving the development of lncRNA biomarkers would potentiate early screening and diagnosis of GDM.
LncRNAs as novel regulators of placental development
Till date there is only one report of genome wide identification of lncRNAs in human term placenta (96). However, there are multiple reports of lncRNAs regulating various functions of placenta during development, primarily focusing on trophoblast cell proliferation (97) and invasion (98). Most of the lncRNAs that were found to regulate trophoblast invasion has been implicated in Preeclampsia and will be discussed in that section of this review.
To identify placental lncRNAs during gestational progress, RNA-seq was performed using human term placenta from male and female fetus (95). A pool of 990 putative and 4,463 known lncRNAs corresponding to 2,899 annotated lncRNA loci was identified. Among them, 1,893 were lincRNAs, 2,012 were antisense lncRNAs, 263 were sense intronic transcripts, and 73 were sense overlapping lncRNAs. Sex-bias comparison revealed differentially expressed lncRNAs HAND2-AS1 and XIST with higher propensities in female libraries as compared to RP1-97J1.2, AC010084.1, and TTTY15 solely restricted to male libraries.
LncRNAs in trophoblast cells
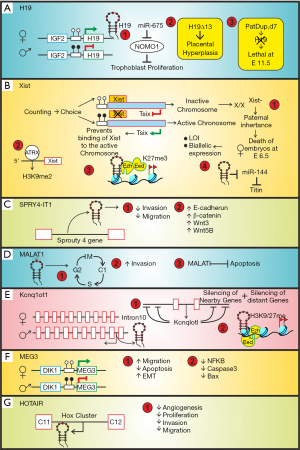
A myriad of lncRNAs still remain to be identified as only a limited number of lncRNAs have been independently analyzed in trophoblast cell types (Figure 3). These include (I) the paternally imprinted H19, (II) X-inactive specific transcript (Xist) and its anti-sense transcript Tsix (III) intronic SPRY4-IT1 (IV) Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) (V) Potassium voltage gated channel, member 1, overlapping transcript 1 (Kcnq1ot1) (VI) Maternally expressed gene 3 (MEG3) and (VII) HOX Transcript AntIsense RNA (HOTAIR).
H19
It was the first lncRNA discovered in pre-genomic era when scientists were using differential hybridization of cDNA libraries to clone and analyze spatiotemporal gene expression in various tissues. At that time it was classified as mRNA but H19 was found to be an unusual transcript that did not translate into protein despite having a small open reading frame. H19 showed high sequence conservation across mammals, transcribed by RNA polymerase II, capped, spliced and polyadenylated and localized to the cytoplasm. Transgenic H19 mice were prenatal lethal suggesting its role in embryonic development as well as importance of its dosage. As a non-coding RNA H19 remained to be mysterious until the functional characterization of another lncRNAs, Xist, which was also found to be involved in dosage compensation in mammals. H19 has been investigated by numerous investigators and was found to be a prototype of multi-tasking lncRNA.
In the developing human placenta, H19 is initially expressed from both paternal and maternal alleles. Interestingly, functional imprinting restricting H19 to the maternal allele starts around 10 weeks of gestation implies that H19 imprinting is established during the development of the placenta (99,100). H19 was reported to be abundant in the RNA from cultured cytotrophoblast cells isolated from human term placenta (101). Interestingly, bi-allelic expression of H19 was found to be exclusive to the extra-villous cytotrophoblasts while the mesenchymal stromal cells retained the mono-allelic expression pattern throughout placental development (102). Mouse embryos harboring a paternal duplication of chromosome 7 (PatDup.d7) lack H19, die at 11.5 dpc and lack spongiotrophoblast (103). H19Δ13 mouse model of overgrowth harboring a deletion of 13kb encompassing the H19 gene and the H19-IGF2 imprinting control region IC1 resulted in 45% increase in placental weight at E19. Owing to loss of imprinting of the adjacent IGF2 gene on the maternal chromosome, these hyperplastic placentas transported 20–35% less glucose and system A amino acids with a 20% reduction in transporter genes like Slc2a3 and Scl38a4 (18,104).
Cai and Cullen [2007] (105) demonstrated that miR675 is encoded by H19 in human keratinocytes. Gao et al. [2012] (106) demonstrated that H19 inhibited human trophoblast cell proliferation via encoding miR-675 that down regulated Nodal Modulator 1 (NOMO1) protein expression. They hypothesized that low expression level of H19 in placenta leading to down regulation of miR-675 that targets NOMO1 and interferes with nodal signaling results in excessive trophoblast cells proliferation and thus marks early-onset severe preeclampsia. Interestingly, forced induction of H19 in mouse ESCs predisposes them to trophoblast lineage commitment and Cdx2 expression even under high Oct3/4 level (107). Functional relevance of H19 lncRNA in trophoblast development is quite evident from the foregoing discussion yet more insights into the mechanism of action remains to be evaluated.
X-inactive specific transcript (Xist) and Tsix
Indispensable function of lncRNA Xist in trophoblast development was first demonstrated by elegant experiments done by Mugford et al. [2012] (108). Lack of any apparent open reading frame in the 17kb transcript of Xist along with its nuclear localization affirmed its categorization as lncRNAs (109). Xist regulates the process of X-chromosome inactivation (XCI) in females. Xist is expressed from and coats the inactivated X chromosome (Xi) in cis. Paternally inherited X chromosome is inactivated by imprinting (methylation) in the female embryos’ extra-embryonic lineage, which gives rise to trophoblast cells, during mouse embryogenesis. Extra-embryonic ectoderm (ExE) cells failed to express CDX2, a marker for trophoblast progenitor cells, at embryonic day 6.5 in female embryos harboring a paternally derived Xist mutation. These embryos subsequently die due to poor trophoblast development. As expected, spongiotrophoblast progenitors, which originate from the CDX2 positive trophoblast stem (TS) cells located within the developing ectoplacental cone, also failed to be maintained. It was further demonstrated that mutation in Xist leads to complete reversal of imprinted XCI patterns in TS cells derived from X/XXist-embryos. These results highlight the importance of Xist mediated dosage compensation for the maintenance of trophoblast progenitors (108). These results were in line with a previous report by Marahrens et al. (110). They showed that transgenic mice with a paternally derived mutation of Xist (X/XXist-) resulted in inheritance dependent female-specific embryonic lethality at E6.5 due to loss of imprinting, biallelic X chromosome expression in the trophoblast and perturbed trophectodermal development. In developing mouse embryo evidence of imprinted XCI is first observed by the two-cell stage (111). “Random XCI” was observed in the epiblast originated from the inner cell mass of the blastocyst. Embryonic lineage reactivates and undergoes a second round of XCI (112), and this time in a “random” way such that paternal and maternal X chromosome have an equal chance of becoming the inactive X (Xi). In contrast, the extra-embryonic cells retain imprinted XCI. It has been demonstrated by Erwin et al. [2012] (113) that mutually exclusive binding of Cdx2 and Oct4 to Xist underlies the switch between imprinted and random XCI.
Genetic and biochemical studies initiated identified numerous proteins in Xist interactome (114-117). Accumulating reports hint towards the role of Xist in epigenetic regulation by chromatin modification (118). Interestingly, the 5’ region of Xist has been identified as a potential nucleation centre for H3K9 dimethylation which serves as a nucleation center for repressive histone and chromatin modifiers (119). At the molecular level, Xist aids in the formation of facultative heterochromatin on the inactive X chromosome by various mechanisms. Firstly, Xist transiently recruit polycomb group protein Eed, Ezh2 and Suz12 to trophoblast giant cells developed from cultured blastocysts thereby initiating H3-K27 methylation (120,121). In addition, ChIP analysis established a stable association of another protein ATRX with the H3K9 dimethylation hotspot 5’ of Xist in an ex vivo model of trophoblast stem (TS) cell differentiation (122). As growing research focus on Xist mediated dosage compensation, regulation of trophoblast specific function by Xist is limited. However, a recent report suggests that XIST potentiates titin expression by inhibiting miR-144 to monitor human trophoblast cell proliferation and invasion (123). To complement the functions of Xist, the expression of Tsix is temporarily regulated to authorize felicitous XCI. Indeed, Tsix assists in methylation of the Xist gene promoter on the maternal allele thereby repressing Xist function and activating the maternal X chromosome.
Murine Xist is positively regulated by Jpx lncRNA and negatively regulated by the antisense Tsix lnc transcript (124-127). Tsix is essential for preventing the inactivation of the maternally inherited X chromosome in extra-embryonic lineages where imprinted X-chromosome inactivation (XCI) occurs. Tsix expression becomes monoallelic and is associated with Xist repression on the active X chromosome (Xa, maternal) at the onset of X inactivation (124). Tsix expression is lost around day 9.5 and Xist repression on the Xa is maintained by other mechanisms, including DNA methylation (128,129). These observations suggest that Tsix regulates the initiation of Xist silencing, but is dispensable for its maintenance.
Unlike in mice, X-inactivation is not imprinted in human placenta and Tsix is not required for XCI (130,131). Due to ethical reasons, XCI is not studied well in early human development. Trans-differentiation of embryonic stem cells to TS cells and chorionic villi hybrid with mouse cells are generally used models for XCI studies. Data on XCI in human placenta point towards random skewed XCI patterns (132).
SPRY4-IT1
SPRY4-IT1 is a 708bp intronic lncRNA encoded from the Sprouty4 gene localized in chromosome 5q31.3 and expressed abundantly in human placenta (133). Expression of this lncRNA increases in preeclamptic placentas (134). In-vitro studies using immortalized first trimester human extravillous cytotrophoblast cells HTR-8/SVneo uncovered the role of this lncRNA in trophoblast cells. SPRY4-IT1 is inhibitory to EMT-associated events like invasion and migration of trophoblast cells. Indeed, the proliferative capacity, and migratory phenotype of HTR-8/SVneo harboring over-expressed SPRY4-IT1 was suppressed as compared to their respective controls. SPTY4-IT1 also induced E-cadherin and β-catenin expression with a concomitant decline in vimentin levels. A reversal of these effects was observed upon down-regulation of SPRY4-IT1 (134,135). RNA immunoprecipitation experiment identified the cytoplasmic RNA binding protein HuR as a novel SPRY4-IT1 interacting protein which actually modulates β-catenin at the post-transcriptional level. Additionally, knockdown of SPRY4-IT1 in HTR-8/SVneo cells diminished WNT3 and WNT5B transcript by 58% and 39% respectively. Altogether, SPRY4-IT1 modulates the canonical Wnt/ β-catenin pathway to inhibit epithelial to mesenchymal transition in trophoblast cells (135). Therefore, SPRY4-IT1 mediated EMT dysfunction of trophoblast cells might serve as a promising therapeutic option to suppress PE.
Metastasis associated lung adenocarcinoma transcript-1 (MALAT-1)
MALAT-1, the first lncRNA associated with human disease was identified in a subtractive hybridization-based screening of transcripts associated with metastasis and patient survival in stage-I non-small cell lung cancer (136). It is a 6.5 kb polyadenylated and unspliced lncRNA localized to human chromosome 11q13 and mouse chromosome 19qA. In contrast to the traditional cleavage and polyadenylation mechanism, the 3’-end of MALAT-1 lacks the poly (A) structure. MALAT-1 over-expression was found in severe forms of invasive placentation, such as placenta accreta (137) prompted investigation on MALAT-1 in trophoblast development. Using first trimester choriocarcinoma cell line, JEG-3, it was demonstrated that shRNA mediated silencing of MALAT-1 leads to suppression of proliferative ability and arrested them in G0/G1 phase of the cell cycle thereby reducing the number of G2/M phase cells. Reduction in MALAT1 expression also triggered the apoptotic pathway and intensified the percentage of apoptotic cells with elevated levels of cleaved caspase-3, cleaved caspase-9 and PARP-1 in addition to impeded migration and invasion in JEG-3 cells (9). Another study suggested that the invasion capability of BeWo, JAR and JEG-3 cells were significantly suppressed after transient knockdown of MALAT-1 (137).
Potassium voltage gated channel, member 1, overlapping transcript 1 (Kcnq1ot1)
Originally named as LIT1, Kcnq1ot1 was identified in a screen of differentially expressed transcripts dictating the genomic organization of the imprinting cluster on human chromosome 11p15 (138). The expression of Kcnq1ot1 begins at the two-cell stage in parallel to Xist and the dynamics of inactivation differs between embryonic and extra-embryonic lineages (139-141). It is a 91kb stable, unspliced, antisense lncRNA expressed from the paternal chromosome (142,143). The transcription start site of Kcnq1ot1 is positioned on the imprinting control region (ICR) located in the intron 10 of KCNQ1 gene which houses 8 other protein-coding genes that expressed from the maternal allele (126). Epigenetic silencing by this lncRNA extends over 400kb in cis in the mouse embryo. Surprisingly, the silenced region spreads over 780kb in the placenta suggesting a critical role of Kcnq1ot1 in placental development.
Similar to Xist, Kcnq1ot1 defines a distinct nuclear domain in cis to epigenetically inactivate genes within the domain boundary. Interestingly, this domain is larger in the placenta as compared to the embryo suggesting a greater number of silenced genes in the placental cluster (143). Although Kcnq1ot1 is transcribed in all tissues, it is only in placenta, where both nearby (Cdkn1c, Kcnq1, Slc22a18, and Phlda2) and distantly placed genes (Cd81, Osbp15, Ascl2, and Tssc4), on either side of the Kcnq1 ICR, are imprinted, whereas embryonic tissues show imprinting of only nearby genes. Interestingly, truncation of the Kcnq1ot1 transcript led to up regulation of all imprinted genes in the domain (144). Kcnq1ot1 RNA, although present at a lower level in placenta, was found to interact with chromatin very strongly in placenta than in other tissues. In placenta, Kcnq1ot1 RNA was found to interact with both PRC2 complex members (Suz12 and Ezh2) and G9a histone methyl transferase (142). In addition, permissible imprinting of the maternally expressed genes by shRNA mediated silencing of Kcnq1ot1 in embryonic and extra-embryonic stem cells specifies that the act of transcription and not post-transcriptional effect of Kcnq1ot1 is critical to early development imprinting maintenance (145).
Maternally expressed gene 3 (MEG3)
Also known as the gene trap locus 2 (Gtl2), Meg3 was identified as a differentially expressed transcript in mouse embryonic development during paternal transmission of lacZ insertion in distal chromosome 12. Despite having multiple ORFs in numerous spliced version, lack of strong Kozak sequence of these ORFs entailed Meg3 gene functions as RNA (146). Later, the human homolog MEG3 was identified as an imprinted gene which is preferentially expressed from the maternal allele (147). Mapped 90kb apart from DLK1 to human chromosome 14q32.3 and mouse distal chromosome 12, MEG3 is the first imprinted gene to be identified at these particular chromosomal loci.
Research initiative undertaken to elucidate Meg3 functionality in placental development is rather limited. The initial clue highlighting Meg3 as a placenta associated lncRNA came from studies exploiting Meg3 knockout mice model carrying a 5.9 kb deletion extending ∼300 bp of the Meg3 promoter till exon five of the gene. Indeed, transmission of the truncated gene from the maternal chromosome emanated in prenatal death of the pups accompanied by silencing of the maternally expressed genes in 18.5 dpc placenta without affecting the growth of the organ (148). Additionally, in vitro data using two different human trophoblast cell lines HTR8/Svneo and JEG-3 suggest that RNA interference-based silencing of endogenous MEG3 impedes migration and enhances apoptosis of trophoblast cells. Also, ectopic over-expression of MEG3 in these cells decreased NFκB, caspase 3 and Bax expression leading to decrease in apoptosis and enhanced their migration (149). In addition, elevated MEG3 also contributes to Smad7 mediated epithelial to mesenchymal transition in HTR8/Svneo by enhancing the expression of mesenchymal bio-markers like N-cadherin, vimentin and slug followed by simultaneous reduction in E-cadherin (150). Epigenetic loss of imprinting at the intergenic MEG3 DMR bestowed “Temple syndrome” in a Caucasian female born to healthy, non-consanguineous parents. Clinical features manifest IUGR, low-birth weight, hypotonia, developmental delay, premature puberty, short stature and truncal obesity (151).
HOTAIR
HOTAIR was identified in a tiling array based characterization of the transcriptional landscape of HOX loci using human adult primary fibroblast (152). HOTAIR is a 2.2 kb lncRNA encoded in the anti-sense direction between HoxC11 and HoxC12 of the HoxC gene cluster on human chromosome 12q13.13. Murine HOTAIR shares about 58% sequence similarity to human HOTAIR (153). Interestingly, the absence of HOTAIR in non-mammalian vertebrates is indicative of its mammalian specific function (154). HOTAIR represents a classical paradigm of trans-acting guide lncRNA.
HOTAIR transcript shows a progressive decrease throughout human gestation. HOTAIR suppresses placental angiogenesis and was found to play a critical role in suppressing the proliferation, invasion, migration and tube formation capacity of HUVECs by functionally down regulating the expression of vascular endothelial growth factor A (97). A lncRNA PCR array identified HOTAIR as a target of YY1 in primary trophoblast cells obtained from first trimester placenta (155). Another report using the same microarray approach highlights that a zinc-finger mRNA binding protein tristetraprolin decreases the half-life of HOTAIR in villous tissue samples (156). A combination of ex vivo trophoblast explants culture with in vitro gain and loss of HOTAIR function in HTR8/Svneo cells identified HOTAIR as an inducer of the invasive potential of trophoblast cells by activating the PI3K/AKT signaling pathway with a surge in MMP2 levels (155). However, a contradictory report using the same cell line highlights HOTAIR as an inhibitor of trophoblast invasion (157). Therefore, additional investigations are necessary to reconfirm the role of HOTAIR in trophoblast biology.
Other lncRNAs associated with trophoblast development and placental biology
In addition to the lncRNAs described above, numerous novel lncRNAs, which regulate placental function, has been reported.
X-linked lncRHOFX1 is expressed robustly in the trophectoderm and primitive endoderm cells of human blastocyst-stage embryos. It is also abundantly expressed in trophoblast progenitors differentiated from human pluripotent stem cells in vitro. It is the first identified lncRNA which provides an excellent concoction of innate immunity dependent viral suppression to early placentation (158).
High expression of lncRNA TERRA (telomeric repeat containing RNA) has been documented in human term placenta. This lncRNA maintains the length of telomeres in human placenta. Promoter hypomethylation in addition to elevated TERRA expression is associated with alternative lengthening of telomeres (ALT) in the absence of hTERT (159).
A novel lncRNA GPR1AS was identified in human GPR1-ZDBF2 intergenic loci on chromosome 2q33.3. This lncRNA displayed placental specific expression, being encoded from the paternal allele. Transcribed in an antisense orientation from introns 2 of GPR1, GPR1AS is devoid of any ZDBF2 exon in the human genome. GPR1AS is a human ortholog of mouse Zdbf2 linc and the epigenetic regulations between GPR1 and ZDBF2 cluster is well conserved between human and mouse (160).
Another study brought forward two long intergenic lncRNAs LINC00629 and MIR503HG between HPRT1 and PLAC1 loci on chromosome Xq26, a region which houses other genes critical to placental development. MIR503HG was exclusively expressed by the placenta and LINC00629 was also highly expressed by placenta and other reproductive tissues like ovary, testis and cervix. Impaired migration and invasion of human choriocarcinoma JEG-3 cells upon ectopic over expression of LINC00629 and MIR503HG highlights its role in human reproduction (161). Another antisense lncRNA GNG12-AS1 mapped to the DIRAS3 locus is biallelically expressed in the placenta (162).
LncRNAs associated with PE
Numerous research works have been initiated to explore global alteration of lncRNAs during the development of PE. Accumulating evidences hint towards equilibrium between pro-proliferative and anti-proliferative lncRNAs as crucial determinants of PE etiologies thereby making lncRNAs potential candidates in PE pathogenesis. Indeed, modulation of trophoblast function and vasculogenic remodelling of the spiral arteries by lncRNAs is imperative to majority of the PE cases observed. Global dynamicity of lncRNA transcripts in PE placentas have been reported by two independent research groups using microarray analysis. The first study involving 32 early onset PE (EOPE) and age-matched preterm control caesarean pregnancies identified 15,646 up-regulated and 13,178 down-regulated lncRNAs as references for future functional studies (163). Another microarray profile exploiting six PE and five normal placenta revealed differential expression of 738 lncRNAs out of which 259 were up regulated and 479 were down regulated in PE cohorts. Validation of this result in 40 PE and control placenta contributed to the fact that dysregulation of three of these highly expressed lncRNAs (LOC391553, LOC284100, CEACAMP8) attributed to PE (164). Other lncRNAs reported in the context of PE pathophysiology include (I) H19, (II) SPRY4-IT1, (III) MALAT1, (IV) MEG3 and (V) HOTAIR.
H19
Loss of the H19 gene imprinting in the placental tissues of pre-eclampsia patients (100,165) and positive regulation of trophoblast invasiveness by H19 prompted interest in role of H19 imprinting in the etiology of PE. This LOI studies were further extended using pyrosequencing-based analysis of global DNA methylation followed by methylation sensitive high resolution melting in PE placentas obtained from a cohort of Chinese patients. This technique demonstrated intensified global DNA methylation along with hypermethylation in the promoter region of H19 with elevated DNMT1 levels followed by a concomitant decrease in H19 transcripts in placentas from early-onset PE (EOPE) patients (106). Likewise, placentas isolated from severe PE (sPE) subjects harboured significant methylation at a single CpG site within exon1 of H19. This observation was further reinforced by treatment of JEG-3 cells with 5-aza-2’-deoxycytidine which reverberated in a surge of H19 transcripts followed by perturbed proliferation, migration and invasion (166). Another study involving 193 PE and 201 control subjects correlated H19 rs217727 single polymorphism to the risk of PE susceptibility (167). Interestingly, a recent study suggested the role of endocrine disrupting chemicals in the imprinting of H19. This pyrosequencing based population study involving 179 pregnant women with 11 phthalate and 8 phenol exposures during the first trimester of pregnancy alters H19 methylation levels resulting in hypomethylation of H19 DMR (168).
SPRY4-IT1
A study involving 25 preeclamptic and normal pregnancy patients suggested 2.8-fold increase in SPRY4-IT1 level in PE placentas as compared to normal ones. In HTR-8/SVneo trophoblast cells SPRY4-IT1 knockdown enhanced the cell migration and proliferation, and reduced the response of cells to apoptosis. As expected, reverse effect was observed by ectopic over expression of SPRY4-IT1 (134). Same group reported three years later that SPRY4-IT1 levels were robustly up regulated in PE placentas as compared to controls in a study involving 50 nulliparous PE and normal pregnancy patients (135). Using HTR-8/SVneo trophoblast cells, it was demonstrated that over expression of SPRY4-IT1 suppressed trophoblast cell migration and invasion, whereas reduced expression of SPRY4-IT1 prevented the epithelial mesenchymal transition (EMT) process. RNA immune-precipitation experiment showed that SPRY4-IT1 bound directly to HuR and altered β-catenin expression in HTR-8/SVneo cells. Furthermore, WNT3 and WNT5B expression were changed following transfection of HTR-8/SVneo with SPRY4-IT1 (135).
Metastasis associated lung adenocarcinoma transcript-1 (MALAT-1)
Only one report has been published so far that highlighted the role of MALAT-1 in trophoblast dysfunction associated with the pathophysiology of PE. A significant reduction in MALAT-1 levels was in preeclamptic placentas as compared to healthy subjects. This data along with in vitro control of MALAT-1 in JEG-3 cells suggests that mitigated MALAT1 might contribute to shallow trophoblast invasion which is a hallmark of PE (98). Lower endogenous MALAT1 levels were reported in mesenchymal stem cells (MSCs) isolated from the umbilical cords of PE patients as compared to healthy donors suggesting impaired apoptosis, enhanced proliferation, migration and invasion and immunosuppressive properties of MSCs in PE (169).
Maternally expressed gene 3 (MEG3)
Decreased proliferation of extra-villous trophoblast cells contributing to incomplete spiral artery remodelling is paramount to PE. Two different studies report significant MEG3 down regulation in placental samples from PE patients as compared to normotensive healthy donors (149,150). MEG 3 is also suggested as a potential candidate regulating vascular smooth muscle cell (VSMC) loss from the maternal uterine spiral arteries by regulating crucial processes of vascular transformation like VSMC migration and apoptosis (170). Overall, depleted MEG3 might potentiate inappropriate spiral artery remodelling in PE subjects which makes it a prime therapeutic target for early intervention of PE.
Hox Transcription Antisense RNA (HOTAIR)
A single report involving 23 severe PE and normotensive patients elucidate that HOTAIR transcript level is increased significantly in placental subjects from PE patients as compared to normal healthy subjects (157). However, additional studies are necessary to delineate the importance of this lncRNA in the pathophysiology of PE. In contrast to HOTAIR, the extent of HOTTIP (a newly discovered lncRNA mapped to the 5’ end of the HoxA cluster on chromosome 7p15.2) is drastically lowered in PE placenta. Reduction of cellular proliferation through G0/G1 arrest by interference of HOTTIP transcript exemplifies synergistic function amidst varied mechanism of two lncRNAs from the same HOX gene but different HOX cluster in the commencement of PE (171).
Other lncRNAs associated with PE
Several novel lncRNAs have been reported to be differentially regulated during the development of PE. Of these, a highly conserved 418bp lncRNA uc.187, 1.7kb RPAIN (transcript variant 12 of RPA interacting protein), CCAT1, DLX-AS1 and lncDC were robustly up regulated in PE placenta (49,172,173). In vitro studies suggested that the proliferative potential and invasiveness of HTR8/Sv-neo cells was enhanced upon silencing uc.187 in accordance with the increase in MMP-2/9, Ki-67 and decrease in TIMP-1. Knockdown of uc.187 inhibited apoptosis with a concomitant down regulation of cleaved caspase-3 and up regulation of Bcl-2 (172). In addition to the anti-proliferative role of RPAIN in HTR8/Sv-neo cells, it modulates trophoblast invasion and apoptosis via the adjacent complement protein C1q thereby providing another therapeutic target for PE diagnosis (174). LncRNA DLX-AS1 is an excellent example of non-coding RNA cross-talk via inhibitory interaction with mir-365c in HTR8/Sv-neo cells to modulate GADD45A in PE (173).
In contrast to the lncRNAs that are over expressed in PE, the level of lnc-ATB, SNHG5, PGK1P2, HK2P1, EGFR-AS1, TUG1, PVT1, linc00473 and MVIH dropped in PE placenta as compared to healthy pregnant biopsies (175-179). Inhibition of endogenous lnc-ATB, PVT1 and TUG1 in HTR8/Sv-neo cells reduced proliferation, migration, invasion and tube formation (175-177). Whereas TUG1 and PVT1 interacted with the polycomb group protein EZH2 to epigenetically silence RND3 and ANGPTL4 transcription respectively, MVIH synergized with Jun-B to modulate trophoblast proliferation (165,176,177). SNHG5 partly controls the miR-26a-5p/N cadherin axis to perturb trophoblast functions in PE (178). Linc00473 particularly interacted with LSD1 to inhibited TFPI2 and EGFR-AS1 inhibited the JAK/STAT pathway during PE progression (179,180). Enthrallingly, PGK1P2 in conjunction with PGK1 and HK2P1 in conjunction with HK2 is imperative to precarious decidualization by acting as competing endogenous RNA (ceRNA) in PE (181,182).
The existence of multiple susceptible loci harboring placentation related genes in PE etiologies is common for decades. Reports from Finnish PE populations exemplify a lncRNA on chromosome 4q35.1 from intron 3 of STOX2 in the regulation of alternative splicing of the gene. Mutation of this lncRNA prematurely truncates the functional protein to introduce a stop codon thereby deleting the C-terminal domain of the STOX2 protein (183).
LncRNAs associated with IUGR
A limited number of lncRNAs have been studied till date in correlation to IUGR pathophysiology. These include (I) H19, (II) Nuclear paraspeckle assembly transcript 1 (NEAT1), and (III) Kcnq1ot1.
H19
Till date H19 is most extensively studied lncRNA in IUGR pathogenesis. Due to their role in cell proliferation, differentiation and trophoblast development, H19/IGF2 imprinted loci prompted investigations on their expression and function in the context of placental disorders. However, the effect of H19 on trophoblast and cancer cell proliferation is rather controversial. H19 has been reported to promote cell proliferation in various cancer types that include colorectal (184), cervical (185), pancreatic ductal (186) carcinoma and many other types of cancers. In contrast, in human trophoblast cells H19 inhibits cell proliferation (106,187). In line with its effect on trophoblast cell proliferation, H19 promoter hypomethylation has been reported in IUGR placentas from normotensive mothers but not in PE (188,189). Zuckerwise et al. [2016] (190) reported contrasting data showing that H19 lncRNA is significantly decreased in placenta from pregnancies with FGR. However, whether these data were from normotensive and hypertensive mothers were not clear from this report. Hypomethylation of the H19 imprinting control region (ICR) in the paternal alleles of the placenta were also reported in ethanol induced mouse model of IUGR (191). In addition, mutation in the telomeric imprinting centre region resulting in biallelic expression of H19 was observed in Silver Russel Syndrome (SRS), a congenital syndrome characterized by severe IUGR (192-194). Nonetheless, the mechanisms by which H19 may impart IUGR phenotype have yet to be elucidated fully.
Nuclear paraspeckle assembly transcript 1 (NEAT1)
NEAT1 was identified using microarray approach to screen for ubiquitously expressed nuclear enriched polyadenylated RNA transcripts in human fibroblast WI-38 and lymphoblastoid cell-line GM00131 cell lines (195). A single report highlights the importance of NEAT1 in the etiology of IUGR. A PCR-select cDNA subtraction using placental biopsies from 12 IUGR and 12 control singleton pregnancy subjects reported a surge in NEAT1 transcript in IUGR patients. In-situ signal of NEAT1 lncRNA was intensified in the nuclei of villous trophoblast cells from IUGR term placenta as compared to normal placentas. Indeed, NEAT1 co-localized with the DBHS protein PSPC1 and the punctate NEAT1 staining is concomitant to the increased number of paraspeckles in IUGR samples Therefore, an upsurge in NEAT1 level would perhaps promote rapid paraspeckle formation to hyperedit mRNAs in IUGR cases (196).
Potassium voltage gated channel, member 1, overlapping transcript 1 (Kcnq1ot1)
Direct reports suggesting the role of Kcnq1ot1 in IUGR is limiting. However, the Kcnq1 imprinting centre houses two growth inhibitory genes CDKN1C and PHLDA2. Indeed, two novel centromere directed cis maternal chromosomal duplication extending 0.88 and 1.13Mb from the middle of Kcnq1 gene respectively display contrasting growth phenotype where 0.88Mb duplication represents the growth retarded phenotype (197).
LncRNAs associated with placental accreta
The only lncRNAs reported to be associated with accrete is Metastasis associated lung adenocarcinoma transcript-1 (MALAT-1). To identify differentially expressed genes, which may impair placentation resulting in placenta previa, increta/percreta (I/P) placental tissues from I/P and non-increta/percreta (non-I/P) sites were concomitantly collected from patients undergoing Cesarean hysterectomy (138). Annealing control primer-polymerase chain reaction (ACP-PCR) followed by cloning and sequencing confirmed over expression of MALAT-1 in I/P samples. Further investigation on the function of MALAT-1 in BeWo, JAR and JEG-3 trophoblast cell lines using siRNA down-regulation led to compromised invasion ability. Thus it was inferred that MALAT-1 directs advanced invasive placentation leading to placenta accreta. However, as discussed earlier that placenta accreta is associated in defective decidualization and in effect decidua basalis is absent in most cases. Therefore, more detailed analysis of origin of placenta accreta and involvement of lncRNAs in pathogenesis remained to be explored.
LncRNAs associated with GDM
Transcriptomic profiling using GeneChip mouse genome microarray in E18 placental samples of GDM mouse model developed by gestational period high-fat diet identified 52 differentially expressed lncRNAs in GDM placentas as compared to control chow fed placentas (198). Hyperglycemic mice model induced by streptozotocin injection however caused H19 hypomethylation resulting in increase H19 transcripts in E10.5 placentas due to adverse uterine environment (199). In GDM patients, the expression level of lncRNA MALAT1 was higher than the controls and correlated with both lncRNA p21 and lncRNA H19 (200). Based on their abundance in GDM, circulating lncRNAs, XLOC_014172 and RP11-230G5.2 have been postulated to act as novel biomarkers in GDM patients as fingerprint for the risk of macrosomia outcome (201).
LncRNAs associated with recurrent miscarriage (RM)
In contrast to infertility, recurrent miscarriage or spontaneous abortion is defined by consecutive loss of pregnancy before 20 completed weeks of gestation. A microarray analysis using chorionic villi tissue samples from 5 RM cases identified 1449 differentially expressed lncRNAs compared to normal pregnancies. Functional analysis of the various pathways regulated by these differentially expressed lncRNAs brought forward ECM-receptor interactions, apoptosis, endocrine and TGF-β signalling as potential deregulated pathways in RM (202). Another attempt exploiting human lncRNA array in decidual samples from sixteen pairs of pregnancies correlated to early spontaneous abortions (SA) and induced abortions (IA) highlighted inflammation and infection pathways as pathophysiological factors underlying SA (203). Interestingly, the level of HOTAIR was reported to be relatively lower in RM trophoblast cells. This together with the in vitro data described earlier makes HOTAIR a potential therapeutic target for RM (155) (Table 2).
Table 2
| Disease | Status | Name of the lncRNAs | References |
|---|---|---|---|
| PE | Down regulated | H19, MALAT1, MEG3, LNC-ATB, TUG1, MVIH, EGFR-AS1, PGK1P2, PVT1, SNHG5, Linc00473 | (98,106,149,150,165,175,177-182) |
| Up regulated | SPRY4-IT1, HOTAIR, Uc.187, RPAIN, DLX6-AS1 | (135,157,172-174) | |
| IUGR | Up regulated | H19, NEAT1 | (190,196) |
| Placenta accreta | Up regulated | MALAT1 | (137) |
| GDM | Up regulated | H19, MALAT1 | (199,200) |
PE, pre-eclampsia; IUGR, intra-uterine growth restriction; GDM, gestational diabetes mellitus; MALAT1, metastasis associated lung adenocarcinoma transcript 1 (MALAT1); NEAT1, nuclear paraspeckle assembly transcript 1; HOTAIR, HOX Transcript AntIsense RNA.
Conclusions
With increasing global scale initiatives to identify non-coding components of the genome, new lncRNAs are being identified and this plethora is likely to be amplified in future. The foregoing discussion provided a survey of the present state of knowledge regarding the discovery, functions, and mechanisms of action of the lncRNAs in context of placental development and disease. The increasing rate of pregnancy associated complications demands major insights towards molecular regulation of placental development and origin of its malfunction leading to devastating pregnancy associated disorders. LncRNAs have gained widespread attention in recent years as potentially new regulators of placental development and function and also been proposed as novel entrants in therapeutics. Although many lncRNAs have been systemically identified, present research is limited to a small range of lncRNAs. Therefore, a large number of lncRNAs and specific mechanism of their action as well as the cross-talk between individual lncRNAs still remains to be explored during the morphogenesis of the placenta.
Acknowledgments
Funding: Supported by CSIR-Indian Institute of Chemical Biology internal support grant. Trishita Basak is a recipient of CSIR pre-doctoral fellowship from the Council of Scientific and Industrial Research, India.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.03.01). RA serves as an unpaid editorial board member of Non-coding RNA Investigation from October 2018 to September 2020. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wen K, Yang L, Xiong T, et al. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res 2016;26:1233-44. [Crossref] [PubMed]
- Chen B, Zhang Y, Zhang X, et al. Genome-wide identification and developmental expression profiling of long noncoding RNAs during Drosophila metamorphosis. Sci Rep 2016;6:23330. [Crossref] [PubMed]
- Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res 2012;22:2529-40. [Crossref] [PubMed]
- Pauli A, Valen E, Lin MF, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 2012;22:577-91. [Crossref] [PubMed]
- Hamazaki N, Uesaka M, Nakashima K, et al. Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development 2015;142:910-20. [Crossref] [PubMed]
- Cui X, Tan J, Shi Y, et al. The long non-coding RNA Gm10768 activates hepatic gluconeogenesis by sequestering microRNA-214 in mice. J Biol Chem 2018;293:4097-109. [Crossref] [PubMed]
- Tang Q, Hann SS. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol Biochem 2018;47:893-913. [Crossref] [PubMed]
- Sathishkumar C, Prabu P, Mohan V, et al. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics 2018;12:41. [Crossref] [PubMed]
- Amit-Avraham I, Pozner G, Eshar S, et al. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A 2015;112:E982-91. [Crossref] [PubMed]
- Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 2012;113:1868-74. [Crossref] [PubMed]
- Sun S, Del Rosario BC, Szanto A, et al. Jpx RNA activates Xist by evicting CTCF. Cell 2013;153:1537-51. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69. [Crossref] [PubMed]
- Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433-45. [Crossref] [PubMed]
- Herriges MJ, Swarr DT, Morley MP, et al. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 2014;28:1363-79. [Crossref] [PubMed]
- Raveendra BL, Swarnkar S, Avchalumov Y, et al. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc Natl Acad Sci U S A 2018;115:E10197-205. [Crossref] [PubMed]
- Massone S, Vassallo I, Fiorino G, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis 2011;41:308-17. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Angiolini E, Coan PM, Sandovici I, et al. Developmental adaptations to increased fetal nutrient demand in mouse genetic models of Igf2-mediated overgrowth. FASEB J 2011;25:1737-45. [Crossref] [PubMed]
- Rosso P. Maternal-fetal exchange during protein malnutrition in the rat. Placental transfer of α-amino isobutyric acid. J Nutr 1977;107:2002-5. [Crossref] [PubMed]
- Red-Horse K, Zhou Y, Genbacev O, et al. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest 2004;114:744-54. [Crossref] [PubMed]
- Aplin JD. Implantation, trophoblast differentiation and haemochorial placentation: mechanistic evidence in vivo and in vitro. J Cell Sci 1991;99:681-92. [PubMed]
- Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice–a review. Placenta 2005;26:S3-9. [Crossref] [PubMed]
- Cross JC. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin Cell Dev Biol 2000;11:105-13. [Crossref] [PubMed]
- Cross JC, Hemberger M, Lu Y, et al. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol Cell Endocrinol 2002;187:207-12. [Crossref] [PubMed]
- Lash GE, Cartwright J, Whitley GSJ, et al. The effects of angiogenic growth factors on extravillous trophoblast invasion and motility. Placenta 1999;20:661-7. [Crossref] [PubMed]
- Iglesias-Platas I, Martin-Trujillo A, Petazzi P, et al. Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum Mol Genet 2014;23:6275-85. [Crossref] [PubMed]
- Maynard SE, Min JY, Merchan J, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest 2003;111:649-58. [Crossref] [PubMed]
- Pardi G, Marconi AM, Cetin I. Placental-fetal interrelationship in IUGR fetuses—a review. Placenta 2002;23:S136-41. [Crossref] [PubMed]
- Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science 2005;308:1592-4. [Crossref] [PubMed]
- Hogg K, Price E, Hanna C, et al. Prenatal and perinatal environmental influences on the human fetal and placental epigenome. Clin Pharmacol Ther 2012;92:716-26. [Crossref] [PubMed]
- Lee SA, Ding C. The dysfunctional placenta epigenome: causes and consequences. Epigenomics 2012;4:561-9. [Crossref] [PubMed]
- Novakovic B, Saffery R. DNA methylation profiling highlights the unique nature of the human placental epigenome. Epigenomics 2010;2:627-38. [Crossref] [PubMed]
- Novakovic B, Saffery R. The ever growing complexity of placental epigenetics–role in adverse pregnancy outcomes and fetal programming. Placenta 2012;33:959-70. [Crossref] [PubMed]
- De Santa F, Barozzi I, Mietton F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010;8:e1000384 [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223. [Crossref] [PubMed]
- Wan Y, Qu K, Zhang QC, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 2014;505:706. [Crossref] [PubMed]
- Cech TR, Steitz JA. The noncoding RNA revolution—trashing old rules to forge new ones. Cell 2014;157:77-94. [Crossref] [PubMed]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet 2015;31:239-51. [Crossref] [PubMed]
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [Crossref] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19:143-57. [Crossref] [PubMed]
- Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev 2015;36:25-64. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621-9. [Crossref] [PubMed]
- Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010;42:1113-7. [Crossref] [PubMed]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009;462:799-802. [Crossref] [PubMed]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest–and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [Crossref] [PubMed]
- Smith KN, Starmer J, Miller SC, et al. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Reports 2017;9:108-21. [Crossref] [PubMed]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 2006;21:617-28. [Crossref] [PubMed]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 2010;143:390-403. [Crossref] [PubMed]
- Venteicher AS, Abreu EB, Meng Z, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009;323:644-8. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Carninci P, Sandelin A, Lenhard B, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006;38:626-35. [Crossref] [PubMed]
- Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002;420:563-73. [Crossref] [PubMed]
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science 2005;309:1564-6. [Crossref] [PubMed]
- Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes. PloS One 2011;6:e25915 [Crossref] [PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915-27. [Crossref] [PubMed]
- Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 2010;28:503-10. [Crossref] [PubMed]
- Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243-50. [Crossref] [PubMed]
- Mercer TR, Gerhardt DJ, Dinger ME, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 2011;30:99. [Crossref] [PubMed]
- Fort A, Hashimoto K, Yamada D, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 2014;46:558-66. [Crossref] [PubMed]
- Seila AC, Calabrese JM, Levine SS, et al. Divergent transcription from active promoters. Science 2008;322:1849-51. [Crossref] [PubMed]
- Sigova AA, Mullen AC, Molinie B, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A 2013;110:2876-81. [Crossref] [PubMed]
- Tani H, Mizutani R, Salam KA, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived non-coding transcripts in mammals. Genome Res 2012;22:947-56. [Crossref] [PubMed]
- Quek XC, Thomson DW, Maag JL, et al. lncRNAdb v2. 0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 2015;43:D168-73. [Crossref] [PubMed]
- Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 2016;44:D203-8. [Crossref] [PubMed]
- Miao YR, Liu W, Zhang Q, et al. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res 2018;46:D276-80. [Crossref] [PubMed]
- Zhi H, Li X, Wang P, Gao Y, et al. Lnc2Meth: a manually curated database of regulatory relationships between long non-coding RNAs and DNA methylation associated with human disease. Nucleic Acids Res 2018;46:D133-8. [Crossref] [PubMed]
- Zhou B, Zhao H, Yu J, et al. EVLncRNAs: a manually curated database for long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res 2018;46:D100-5. [Crossref] [PubMed]
- Salhi A, Essack M, Alam T, et al. DES-ncRNA: A knowledgebase for exploring information about human micro and long noncoding RNAs based on literature-mining. RNA Biol 2017;14:963-71. [Crossref] [PubMed]
- Mas-Ponte D, Carlevaro-Fita J, Palumbo E, et al. LncATLAS database for subcellular localisation of long noncoding RNAs. RNA 2017;23:1080-7. [Crossref] [PubMed]
- Alam T, Uludag M, Essack M, et al. FARNA: knowledgebase of inferred functions of non-coding RNA transcripts. Nucleic Acids Res 2017;45:2838-48. [PubMed]
- Ning S, Zhao Z, Ye J, et al. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinformatics 2014;15:152. [Crossref] [PubMed]
- Chen X, Hao Y, Cui Y, et al. LncVar: a database of genetic variation associated with long non-coding genes. Bioinformatics 2017;33:112-8. [Crossref] [PubMed]
- Hao Y, Wu W, Li H, et al. NPInter v3. 0: an upgraded database of noncoding RNA-associated interactions. Database 2016;2016:baw057 [Crossref] [PubMed]
- Zheng LL, Li JH, Wu J, et al. deepBase v2. 0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 2016;44:D196-202. [Crossref] [PubMed]
- Zhou Z, Shen Y, Khan MR, et al. LncReg: a reference resource for lncRNA-associated regulatory networks. Database 2015;2015:bav083 [Crossref] [PubMed]
- Volders PJ, Verheggen K, Menschaert G, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res 2015;43:D174-80. [Crossref] [PubMed]
- Jiang Q, Wang J, Wu X, et al. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res 2015;43:D193-6. [Crossref] [PubMed]
- Bhartiya D, Pal K, Ghosh S, et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database 2013;2013:bat034 [Crossref] [PubMed]
- Das S, Ghosal S, Sen R, Chakrabarti J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PloS One 2014;9:e98965 [Crossref] [PubMed]
- Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35:W345-9 [Crossref] [PubMed]
- Hu L, Xu Z, Hu B, et al. COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res 2017;45:e2 [Crossref] [PubMed]
- Wang L, Park HJ, Dasari S, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 2013;41:e74 [Crossref] [PubMed]
- Yi X, Zhang Z, Ling Y, et al. PNRD: a plant non-coding RNA database. Nucleic Acids Res 2015;43:D982-9. [Crossref] [PubMed]
- Boyd J, Hamilton W. The human placenta. Cambridge: W. Heffer and Sons. 1970;8:77-8.
- Moore KL, Persaud T. The Developing Human: Clinically Oriented Embryology. Saunders 2004.
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet 2001;2:538-48. [Crossref] [PubMed]
- Vijayaselvi R, Cherian AG. Risk assessment of intrauterine growth restriction. Curr Med Issues 2017;15:262-6. [Crossref]
- Lauenborg J, Mathiesen E, Hansen T, et al. The prevalence of the metabolic syndrome in a Danish population of women with previous gestational diabetes mellitus is three-fold higher than in the general population. J Clin Endocrinol Metab 2005;90:4004-10. [Crossref] [PubMed]
- Shah BR, Retnakaran R, Booth GL. Increased risk of cardiovascular disease in young women following gestational diabetes mellitus. Diabetes Care 2008;31:1668-9. [Crossref] [PubMed]
- Zhu WW, Yang HX, Wei YM, et al. Evaluation of the value of fasting plasma glucose in the first prenatal visit to diagnose gestational diabetes mellitus in China. Diabetes Care 2013;36:586-90. [Crossref] [PubMed]
- Hod M, Kapur A, Sacks DA, et al. The International Federation of Gynecology and Obstetrics (FIGO) Initiative on gestational diabetes mellitus: A pragmatic guide for diagnosis, management, and care. Int J Gynecol Obstet 2015;131:S173-211. [Crossref]
- Syngelaki A, Kotecha R, Pastides A, et al. First-trimester biochemical markers of placentation in screening for gestational diabetes mellitus. Metabolism 2015;64:1485-9. [Crossref] [PubMed]
- Farina A, Eklund E, Bernabini D, et al. A first-trimester biomarker panel for predicting the development of gestational diabetes. Reprod Sci 2017;24:954-9. [Crossref] [PubMed]
- Majewska M, Lipka A, Paukszto L, et al. Preliminary RNA-Seq Analysis of Long Non-Coding RNAs Expressed in Human Term Placenta. Int J Mol Sci 2018;19:1894. [Crossref] [PubMed]
- Wu K, Liu F, Wu W, et al. Long non-coding RNA HOX transcript antisense RNA (HOTAIR) suppresses the angiogenesis of human placentation by inhibiting vascular endothelial growth factor A expression. Reprod Fertil Dev 2018; [Epub ahead of print]. [PubMed]
- Chen H, Meng T, Liu X, et al. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int J Clin Exp Pathol 2015;8:12718-27. [PubMed]
- Jinno Y, Ikeda Y, Yun K, et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat Genet 1995;10:318-24. [Crossref] [PubMed]
- Yu L, Chen M, Zhao D, et al. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta 2009;30:443-7. [Crossref] [PubMed]
- Rachmilewitz J, Gileadi O, Eldar-Geva T, et al. Transcription of the H19 gene in differentiating cytotrophoblasts from human placenta. Mol Reprod Dev 1992;32:196-202. [Crossref] [PubMed]
- Adam GI, Cui H, Miller SJ, et al. Allele-specific in situ hybridization (ASISH) analysis: a novel technique which resolves differential allelic usage of H19 within the same cell lineage during human placental development. Development 1996;122:839-47. [PubMed]
- McLaughlin KJ, Szabó P, Haegel H, Mann JR. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development 1996;122:265-70. [PubMed]
- Esquiliano DR, Guo W, Liang L, et al. Placental glycogen stores are increased in mice with H19 null mutations but not in those with insulin or IGF type 1 receptor mutations. Placenta 2009;30:693-9. [Crossref] [PubMed]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007;13:313-6. [Crossref] [PubMed]
- Gao WL, Liu M, Yang Y, et al. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol 2012;9:1002-10. [Crossref] [PubMed]
- Fujimori H, Mukai H, Murakami Y, et al. The H19 induction triggers trophoblast lineage commitment in mouse ES cells. Biochem Biophys Res Commun 2013;436:313-8. [Crossref] [PubMed]
- Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Development 2012;139:2130-8. [Crossref] [PubMed]
- Clemson CM, McNeil JA, Willard HF, et al. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 1996;132:259-75. [Crossref] [PubMed]
- Marahrens Y, Panning B, Dausman J, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 1997;11:156-66. [Crossref] [PubMed]
- Namekawa SH, Payer B, Huynh KD, et al. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol 2010;30:3187-205. [Crossref] [PubMed]
- Tan SS, Williams EA, Tam PP. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet 1993;3:170-4. [Crossref] [PubMed]
- Erwin JA, del Rosario B, Payer B, et al. An ex vivo model for imprinting: mutually exclusive binding of Cdx2 and Oct4 as a switch for imprinted and random X-inactivation. Genetics 2012;192:857-68. [Crossref] [PubMed]
- Chu C, Zhang QC, Da Rocha ST, et al. Systematic discovery of Xist RNA binding proteins. Cell 2015;161:404-16. [Crossref] [PubMed]
- McHugh CA, Chen CK, Chow A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015;521:232-6. [Crossref] [PubMed]
- Minajigi A, Froberg JE, Wei C, et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015;349:aab2276 [Crossref] [PubMed]
- Monfort A, Di Minin G, Postlmayr A, et al. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep 2015;12:554-61. [Crossref] [PubMed]
- Fang J, Chen T, Chadwick B, et al. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 2004;279:52812-5. [Crossref] [PubMed]
- Rougeulle C, Chaumeil J, Sarma K, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol 2004;24:5475-84. [Crossref] [PubMed]
- Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003;300:131-5. [Crossref] [PubMed]
- de la Cruz CC, Fang J, Plath K, et al. Developmental regulation of Suz12 localization. Chromosoma 2005;114:183-92. [Crossref] [PubMed]
- Baumann C, De La Fuente R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma 2009;118:209-22. [Crossref] [PubMed]
- Yu N, Liang Y, Zhu H, et al. CsA Promotes XIST Expression to Regulate Human Trophoblast Cells Proliferation and Invasion Through miR-144/Titin Axis. J Cell Biochem 2017;118:2208-18. [Crossref] [PubMed]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 1999;21:400-4. [Crossref] [PubMed]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 2000;103:17-27. [Crossref] [PubMed]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 1999;99:47-57. [Crossref] [PubMed]
- Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci U S A 2001;98:10232-7. [Crossref] [PubMed]
- Barr H, Hermann A, Berger J, et al. Mbd2 contributes to DNA methylation-directed repression of the Xist gene. Mol Cell Biol 2007;27:3750-7. [Crossref] [PubMed]
- Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev 1995;9:2325-34. [Crossref] [PubMed]
- Migeon BR, Chowdhury AK, Dunston JA, et al. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am J Hum Genet 2001;69:951-60. [Crossref] [PubMed]
- Migeon BR, Lee CH, Chowdhury AK, et al. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am J Hum Genet 2002;71:286-93. [Crossref] [PubMed]
- van den Berg IM, Galjaard R, Laven J, et al. XCI in preimplantation mouse and human embryos: first there is remodelling. Hum Genet 2011;130:203-15. [Crossref] [PubMed]
- Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 2011;71:3852-62. [Crossref] [PubMed]
- Zou Y, Jiang Z, Yu X, et al. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PloS One 2013;8:e79598 [Crossref] [PubMed]
- Zuo Q, Huang S, Zou Y, et al. The Lnc RNA SPRY4-IT1 modulates trophoblast cell invasion and migration by affecting the epithelial-mesenchymal transition. Sci Rep 2016;6:37183. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Tseng JJ, Hsieh YT, Hsu SL, et al. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod 2009;15:725-31. [Crossref] [PubMed]
- Mitsuya K, Meguro M, Lee MP, et al. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 1999;8:1209-17. [Crossref] [PubMed]
- Green K, Lewis A, Dawson C, et al. A developmental window of opportunity for imprinted gene silencing mediated by DNA methylation and the Kcnq1ot1 noncoding RNA. Mamm Genome 2007;18:32-42. [Crossref] [PubMed]
- Lewis A, Green K, Dawson C, et al. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development 2006;133:4203-10. [Crossref] [PubMed]
- Okamoto I, Otte AP, Allis CD, et al. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 2004;303:644-9. [Crossref] [PubMed]
- Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008;32:232-46. [Crossref] [PubMed]
- Redrup L, Branco MR, Perdeaux ER, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 2009;136:525-30. [Crossref] [PubMed]
- Mancini-Dinardo D, Steele SJ, Levorse JM, et al. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 2006;20:1268-82. [Crossref] [PubMed]
- Golding MC, Magri LS, Zhang L, et al. Depletion of Kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development 2011;138:3667-78. [Crossref] [PubMed]
- Schuster-Gossler K, Bilinski P, Sado T, et al. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn 1998;212:214-28. [Crossref] [PubMed]
- Miyoshi N, Wagatsuma H, Wakana S, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000;5:211-20. [Crossref] [PubMed]
- Zhou Y, Cheunsuchon P, Nakayama Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 2010;137:2643-52. [Crossref] [PubMed]
- Zhang Y, Zou Y, Wang W, et al. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J Cell Biochem 2015;116:542-50. [Crossref] [PubMed]
- Yu L, Kuang LY, He F, et al. The Role and Molecular Mechanism of Long Nocoding RNA-MEG3 in the Pathogenesis of Preeclampsia. Reprod Sci 2018;25:1619-28. [Crossref] [PubMed]
- Briggs TA, Lokulo-Sodipe K, Chandler KE, et al. Temple syndrome as a result of isolated hypomethylation of the 14q32 imprinted DLK1/MEG3 region. Am J Med Genet A 2016;170A:170-5. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 2011;7:e1002071 [Crossref] [PubMed]
- Wu Y, Zhang L, Wang Y, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol 2014;35:9531-8. [Crossref] [PubMed]
- Zhang Y, Jin F, Li XC, et al. The YY1-HOTAIR-MMP2 signaling axis controls trophoblast invasion at the maternal-fetal interface. Mol Ther 2017;25:2394-403. [Crossref] [PubMed]
- Tian FJ, He XY, Wang J, et al. Elevated Tristetraprolin Impairs Trophoblast Invasion in Women with Recurrent Miscarriage by Destabilization of HOTAIR. Mol Ther Nucleic Acids 2018;12:600-9. [Crossref] [PubMed]
- Zou YF, Sun L. Long noncoding RNA HOTAIR modulates the function of trophoblast cells in pre-eclampsia. Sichuan Da Xue Xue Bao Yi Xue Ban 2015;46:113-7, 122. [PubMed]
- Penkala I, Wang J, Syrett CM, et al. LNCRHOXF1: a long noncoding RNA from the X-chromosome that suppresses viral response genes during development of the early human placenta. Mol Cell Biol 2016;36:1764-75. [Crossref] [PubMed]
- Novakovic B, Napier CE, Vryer R, et al. DNA methylation mediated up-regulation of TERRA non-coding RNA is coincident with elongated telomeres in the human placenta. Mol Hum Reprod 2016;22:791-9. [Crossref] [PubMed]
- Kobayashi H, Yanagisawa E, Sakashita A, et al. Epigenetic and transcriptional features of the novel human imprinted lncRNA GPR1AS suggest it is a functional ortholog to mouse Zdbf2linc. Epigenetics 2013;8:635-45. [Crossref] [PubMed]
- Muys BR, Lorenzi JCC, Zanette DL, et al. Placenta-enriched lincRNAs MIR503HG and LINC00629 decrease migration and invasion potential of JEG-3 cell line. PLoS One 2016;11:e0151560 [Crossref] [PubMed]
- Niemczyk M, Ito Y, Huddleston J, et al. Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA. Am J Hum Genet 2013;93:224-35. [Crossref] [PubMed]
- Long W, Rui C, Song X, et al. Distinct expression profiles of lncRNAs between early-onset preeclampsia and preterm controls. Clin Chim Acta 2016;463:193-9. [Crossref] [PubMed]
- He X, He Y, Xi B, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One 2013;8:e81437 [Crossref] [PubMed]
- Zhao D, Li L, Yu L, et al. H19 expression in placenta with pre-eclampsia. Zhonghua Fu Chan Ke Za Zhi 2009;44:87-90. [PubMed]
- Lu L, Hou Z, Li L, et al. Methylation pattern of H19 exon 1 is closely related to preeclampsia and trophoblast abnormalities. Int J Mol Med 2014;34:765-71. [Crossref] [PubMed]
- Harati-Sadegh M, Kohan L, Teimoori B, et al. The long non-coding RNA H19 rs217727 polymorphism is associated with PE susceptibility. J Cell Biochem 2018;119:5473-80. [Crossref] [PubMed]
- LaRocca J, Binder AM, McElrath TF, et al. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res 2014;133:396-406. [Crossref] [PubMed]
- Li X, Song Y, Liu F, et al. Long non-coding RNA MALAT1 promotes proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing VEGF and IDO. J Cell Biochem 2017;118:2780-91. [Crossref] [PubMed]
- Liu W, Liu X, Luo M, et al. dNK derived IFN-γ mediates VSMC migration and apoptosis via the induction of LncRNA MEG3: a role in uterovascular transformation. Placenta 2017;50:32-9. [Crossref] [PubMed]
- Li L, Zhang X, Hong S, et al. Long non-coding HOTTIP regulates preeclampsia by inhibiting RND3. Eur Rev Med Pharmacol Sci 2018;22:3277-85. [PubMed]
- Cao C, Li J, Li J, et al. Long Non-Coding RNA Uc. 187 Is Upregulated in Preeclampsia and Modulates Proliferation, Apoptosis, and Invasion of HTR-8/SVneo Trophoblast Cells. J Cell Biochem 2017;118:1462-70. [Crossref] [PubMed]
- Tan Y, Xiao D, Xu Y, et al. Long non-coding RNA DLX6-AS1 is upregulated in preeclampsia and modulates migration and invasion of trophoblasts through the miR-376c/GADD45A axis. Exp Cell Res 2018;370:718-24. [Crossref] [PubMed]
- Song X, Rui C, Meng L, et al. Long non-coding RNA RPAIN regulates the invasion and apoptosis of trophoblast cell lines via complement protein C1q. Oncotarget 2017;8:7637-46. [PubMed]
- Liu X, Chen H, Kong W, et al. Down-regulated long non-coding RNA-ATB in preeclampsia and its effect on suppressing migration, proliferation, and tube formation of trophoblast cells. Placenta 2017;49:80-7. [Crossref] [PubMed]
- Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis 2017;8:e3104 [Crossref] [PubMed]
- Xu Y, Lian Y, Zhang Y, et al. The long non-coding RNA PVT 1 represses ANGPTL 4 transcription through binding with EZH 2 in trophoblast cell. J Cell Mol Med 2018;22:1272-82. [PubMed]
- Yang Y, Xi L, Ma Y, et al. The lncRNA small nucleolar RNA host gene 5 regulates trophoblast cell proliferation, invasion, and migration via modulating miR-26a-5p/N-cadherin axis. J Cell Biochem 2019;120:3173-84. [Crossref] [PubMed]
- Zhao ZM, Jiang J. Lowly expressed EGFR-AS1 promotes the progression of preeclampsia by inhibiting the EGFR-JAK/STAT signaling pathway. Eur Rev Med Pharmacol Sci 2018;22:6190-7. [PubMed]
- Wu D, Xu Y, Zou Y, et al. Long Noncoding RNA 00473 Is Involved in Preeclampsia by LSD1 Binding-Regulated TFPI2 Transcription in Trophoblast Cells. Mol Ther Nucleic Acids 2018;12:381-92. [Crossref] [PubMed]
- Lv H, Tong J, Yang J, et al. Dysregulated Pseudogene HK2P1 May Contribute to Preeclampsia as a Competing Endogenous RNA for Hexokinase 2 by Impairing Decidualization. Hypertension. 2018;71:648-58. [Crossref] [PubMed]
- Tong J, Yang J, Lv H, et al. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens 2018;13:37-45. [Crossref] [PubMed]
- Oudejans CB, Poutsma A, Michel OJ, et al. Noncoding RNA-regulated gain-of-function of STOX2 in Finnish pre-eclamptic families. Sci Rep 2016;6:32129. [Crossref] [PubMed]
- Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing β-catenin expression in colorectal cancer. Biomed Res Int 2017;2017:2767484 [PubMed]
- Ou L, Wang D, Zhang H, et al. Decreased expression of MiR-138-5p by LncRNA H19 in cervical cancer promotes tumor proliferation. Oncol Res 2018;26:401-10. [Crossref] [PubMed]
- Ma L, Tian X, Wang F, et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther 2016;17:1051-61. [Crossref] [PubMed]
- Yu LL, Chang K, Lu LS, et al. Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC Cell Biol 2013;14:26. [Crossref] [PubMed]
- Bourque DK, Avila L, Penaherrera M, et al. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197-202. [Crossref] [PubMed]
- Tabano S, Colapietro P, Cetin I, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics 2010;5:313-24. [Crossref] [PubMed]
- Zuckerwise L, Li J, Lu L, et al. H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TβR3 in placentae with fetal growth restriction. Oncotarget 2016;7:38398-407. [Crossref] [PubMed]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol Reprod 2009;81:618-27. [Crossref] [PubMed]
- Gicquel C, Rossignol S, Cabrol S, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet 2005;37:1003-7. [Crossref] [PubMed]
- Lee DH, Singh P, Tsark WM, et al. Complete biallelic insulation at the H19/Igf2 imprinting control region position results in fetal growth retardation and perinatal lethality. PloS One 2010;5:e12630 [Crossref] [PubMed]
- Yamazawa K, Kagami M, Fukami M, et al. Monozygotic female twins discordant for Silver-Russell syndrome and hypomethylation of the H19-DMR. J Hum Genet 2008;53:950-5. [Crossref] [PubMed]
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [Crossref] [PubMed]
- Gremlich S, Damnon F, Reymondin D, et al. The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development. Placenta 2014;35:44-9. [Crossref] [PubMed]
- Boonen SE, Freschi A, Christensen R, et al. Two maternal duplications involving the CDKN1C gene are associated with contrasting growth phenotypes. Clin Epigenetics 2016;8:69. [Crossref] [PubMed]
- Huang C, Huang BB, Niu JM, et al. Global mRNA and Long Non-Coding RNA Expression in the Placenta and White Adipose Tissue of Mice Fed a High-Fat Diet During Pregnancy. Cell Physiol Biochem 2018;50:2260-71. [Crossref] [PubMed]
- Ge ZJ, Liang QX, Luo SM, et al. Diabetic uterus environment may play a key role in alterations of DNA methylation of several imprinted genes at mid-gestation in mice. Reprod Biol Endocrinol 2013;11:119. [Crossref] [PubMed]
- Zhang Y, Wu H, Wang F, et al. Long non-coding RNA MALAT1 expression in patients with gestational diabetes mellitus. Int J Gynecol Obstet 2018;140:164-9. [Crossref] [PubMed]
- Lu J, Wu J, Zhao Z, et al. Circulating LncRNA Serve as Fingerprint for Gestational Diabetes Mellitus Associated with Risk of Macrosomia. Cell Physiol Biochem 2018;48:1012-8. [Crossref] [PubMed]
- Wang L, Tang H, Xiong Y, et al. Differential expression profile of long noncoding RNAs in human chorionic villi of early recurrent miscarriage. Clin Chim Acta 2017;464:17-23. [Crossref] [PubMed]
- Wang H, Cao Q, Ge J, et al. Lnc RNA-regulated Infection and Inflammation Pathways Associated with Pregnancy Loss: Genome Wide Differential Expression of lnc RNA s in Early Spontaneous Abortion. Am J Reprod Immunol 2014;72:359-75. [Crossref] [PubMed]
Cite this article as: Basak T, Ain R. Long non-coding RNAs in placental development and disease. Non-coding RNA Investig 2019;3:14.


