
Target RNA-directed microRNA degradation; which controls which?
In general, microRNAs (miRNAs) control target gene expression by inhibiting translation and destroying target RNAs (1,2). However, interestingly some endogenous coding or noncoding RNAs have a function to sequester miRNAs from the real target RNAs by hybridization (called sponge function) without degradation and they are also known as competing endogenous RNAs (ceRNAs) (3,4). On the other hand, although miRNAs are usually stable, accumulating evidence suggests that endogenous transcripts or sometimes viral transcripts also control miRNAs by target RNA-directed miRNA degradation (TDMD) (5-7). In this degradation process, the complementarity between miRNA and target RNA is very critical (Figure 1) (7). miRNAs are quite stable in RNA-induced silencing complex (RISC) if the pairing with target RNA is partial (Figure 1A), although the extensive pairing with target RNA induces 3’ trimming, tailing and degradation of miRNA (Figure 1B). Therefore, some potential target RNAs of miRNAs are not simply targets of miRNAs but also control miRNA’s function and stability. Despite unclear significance in other systems and mechanisms of TDMD before, recent reports have demonstrated the strong evidence and mechanisms of TDMD.
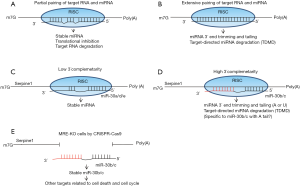
Ghini et al. reported more detailed mechanisms of TDMD of miR-30 family (8). The authors initially classified miRNA: target pairs (TargetScan 6.2) into three groups (high, mild and low) based on 3’ complementarity. Only 6% of pairs are categorized in the high group. Interestingly, in the members of miR-30, miR-30b and miR-30c (miR-30b/c) have higher 3’ complementarity to target site in Serpine1 3’UTR but miR-30a/d/e have the lower complementarity (Figure 1C,D). miR-30b/c is downregulated very fast in the serum depletion and re-fed experiments, indicating that miR-30b/c is more sensitive to TDMD than miR-30a/d/e. miR-30b/c is more efficiently subjected to 3’ end trimming and tailing (A, adenylation; or U, uridylation) and degradation (Figure 1D). The authors also suggested that TDMD is more effective at high ratios (>10) of target RNA over miRNA. Because the complementarity to target site in Serpine1 3’UTR is critical, the authors created cells with deletion of miRNA responsive element (MRE) in Serpine1 3’UTR [MRE-knockout (KO)] using the technique of CRISPR-Cas9 system (Figure 1E). miR-30b/c is more stable in MRE-KO cells compared to wild type (WT) cells. More interestingly, A-forms (with A tail) of miR-30b/c are significantly decreased in MRE-KO cells, suggesting that A-forms but not U-form of miR-30b/c are efficiently subjected to TDMD. As well as detailed mechanisms of TDMD, gene expression was compared between WT and MRE-KO cells. Apoptosis and cell cycle-related genes are enriched in differentially expressed genes between WT and MRE-KO cells and correlated with the cell phenotypes (cell death, cell cycle and proliferation). Those results suggest that miR-30b/c escaped from TDMD in MRE-KO cells target other miR-30 target RNAs (cell death and cycle-related genes) and affect the cell cycle and death.
A recent report showed that the 3’UTR of neuronal-regeneration-related protein (NREP) has near-perfect miR-29b site and destabilize miR-29b through 3’ trimming in brain that affects behavior of mice and Zebra fishes (9). In another report, a long noncoding RNA Cyrano hybridizes and triggers miR-7 degradation in the brain to control neuronal activity also through a circular RNA (Cdr1) and another miRNA miR-671 in the very sophisticated network (10). Those recent findings have demonstrated that TDMD is involved in brain function and behavior of animals but not only in cell death and cell cycle regulation. Therefore, depending on the conditions (3’ complementarity and ratio of miRNA: target RNA), miRNAs control target RNAs but target RNAs can also control miRNA stability (TDMD). This bidirectional regulation may be very important for fine tuning to balance the gene expression or avoid extremely high or low expression.
For the therapeutics targeting miRNAs, perfect-matched antisense oligonucleotides (ASOs) have been used to destabilize target miRNAs in several cases (11-15) although the detailed mechanisms are not very clear. But trimming of 3’ end and degradation of miRNAs hybridized with perfect-matched ASO (possibly TDMD in RISC) may be a mechanism of suppression of miRNA function (degradation) although RNase H-dependent degradation of target RNAs in RNA/DNA duplexes could also be another mechanism (16). Utilizing those natural systems (TDMD or RNase H-mediated degradation) may be a wiser way to develop ASO therapeutics to treat human diseases in the pathogenesis that miRNAs are involved. TDMD may provide a new logical design for ASO targeting non-coding RNAs including miRNAs in potential therapeutics (17-21). Understanding the detailed mechanisms of TDMD would provide not only fundamental and basic discoveries but also new therapeutics for human diseases.
Acknowledgments
Funding: The author is grateful to the funding from the National Institutes of Health, the Wanek Family Project to Cure Type 1 Diabetes at City of Hope and the Juvenile Diabetes Research Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Jing Shi (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.11.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bartel DP. Metazoan MicroRNAs. Cell 2018;173:20-51. [Crossref] [PubMed]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015;16:421. [Crossref] [PubMed]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Ameres SL, Horwich MD, Hung JH, et al. Target RNA–Directed Trimming and Tailing of Small Silencing RNAs. Science 2010;328:1534-9. [Crossref] [PubMed]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010;328:1563-6. [Crossref] [PubMed]
- Pasquinelli AE. Paring MiRNAs Through Pairing. Science 2010;328:1494-5. [Crossref] [PubMed]
- Ghini F, Rubolino C, Climent M, et al. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat Commun 2018;9:3119. [Crossref] [PubMed]
- Bitetti A, Mallory AC, Golini E, et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 2018;25:244-51. [Crossref] [PubMed]
- Kleaveland B, Shi CY, Stefano J, et al. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018;174:350-62.e17. [Crossref] [PubMed]
- Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87-98. [Crossref] [PubMed]
- Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881-9. [Crossref] [PubMed]
- Putta S, Lanting L, Sun G, et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 2012;23:458-69. [Crossref] [PubMed]
- Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010;28:341. [Crossref] [PubMed]
- Kato M. Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease. Kidney Res Clin Pract 2018;37:197-209. [Crossref] [PubMed]
- Frieden M, Christensen SM, Mikkelsen ND, et al. Expanding the design horizon of antisense oligonucleotides with alpha–l–LNA. Nucleic Acids Research 2003;31:6365-72. [Crossref] [PubMed]
- Crooke ST, Witztum JL, Bennett CF, et al. RNA-Targeted Therapeutics. Cell Metabolism 2018;27:714-39. [Crossref] [PubMed]
- Kato M, Wang M, Chen Z, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 2016;7:12864. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Kato M, Natarajan R. Diabetic nephropathy[mdash]emerging epigenetic mechanisms. Nat Rev Nephrol 2014;10:517-30. [Crossref] [PubMed]
- Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 2015;11:23-33. [Crossref] [PubMed]
Cite this article as: Kato M. Target RNA-directed microRNA degradation; which controls which? Non-coding RNA Investig 2018;2:62.


