
The parasitic plant haustorium: a trojan horse releasing microRNAs that take control of the defense responses of the host
Organisms in a symbiotic relationship have evolved in an arms race to modulate each other’s physiology. In most cases, symbiotic interactions elicit a local and a systemic response that involves an extensive transcriptional reprogramming of both organisms. Although the genetic component of these interactions has been studied for years, the potential exchange of genetic information between symbionts has remained elusive till now. The theory that an exchange of RNA could exist was an exciting, yet difficult-to-prove, theory. However, in recent years, due to both the development of high-throughput sequencing and increase in genome sequence information, it has been confirmed that both messenger RNAs (mRNAs) and small RNAs (sRNAs) are exchanged between symbionts. Most strikingly, these foreign sRNAs are functional in the host cellular environment and, in most of the cases, they are involved in the regulation of pathogenicity and virulence. Here, we review the recent discovery of the exchange of sRNAs between a parasitic plant, Cuscuta campestris, and its host Arabidopsis thaliana (1), which opens new perspectives and questions both at the agricultural and biological levels.
RNA silencing in plants
RNAi is an evolutionary conserved mechanism in eukaryotes that has diversified particularly in plants, where it is termed RNA silencing (2). The roles of RNA silencing range from RNA degradation to promotion of cytosine methylation in the genomic DNA. Together, these events lead to the silencing of the gene or genomic region complementary to the sRNA at the transcriptional or post-transcriptional levels. In broad terms, the initial event of the mechanism is the processing of a double stranded RNA (dsRNA) by Dicer-like (DCL) proteins into sRNAs of 20–24 nts in size, which are the hallmark of the mechanism. These sRNAs are later loaded into Argonaute (AGO) proteins that form the RNA-induced silencing complex (RISC) that scans for other RNAs with complementarity to the sRNA and exert the function of the AGO protein. The sequence specificity mediated by the sRNA is another of the hallmarks of all these pathways (3).
The various functions that the different RNA silencing pathways accomplish include defense against exogenous RNAs (like viruses or transgenes) by the production of phased small-interfering RNA (phasiRNAs) (4), coordination of the developmental signals by the microRNA (miRNA) pathway (5) and control of genome stability through the regulation of transposable elements and promotion of heterochromatin by the RNA-directed DNA methylation pathway (6). The miRNA pathway is involved in the control of the expression of several genes that regulate plant development including multiple transcription factors like APETALA2 or Scarecrow-like (7). miRNAs are sRNAs of 21 nts in length produced from non-coding transcripts with high levels of self-complementarity that have very characteristic hairpin-like secondary RNA structures. These RNA hairpins are processed by DCL1 resulting in the production of the miRNA sequences, which are very stable and evolutionary conserved. miRNAs are then loaded into AGO1, AGO7 or AGO10, which induce the cleavage or translational repression of their target mRNAs (3). phasiRNAs are 21–22 nt long sRNAs that are produced from coding or non-coding transcripts that were marked for degradation by specific 22 nt long miRNAs. This process is characterized by the biogenesis of a dsRNA mediated by the RNA-directed RNA polymerase 6 (RDR6) that is subsequently processed by DCL4 and DCL2 into 21 and 22 nt siRNAs that are generated “in phase”. Once produced, phasiRNAs can target their original transcript, initiating an amplification of the original silencing signal or can target genes that have a high degree of similarity with the original transcript. This is the case for the regulation of TAS genes in Arabidopsis or leucine-rich repeat genes (NB-LRR) in multiple species (8).
Trans-species mobility of sRNAs
The observation that RNA silencing produces mobile signals that maintain the silencing status at distant tissues was evident from almost the first observations of the RNA silencing phenomenon (9,10). Indeed, sRNAs can move both from cell to cell and over long distances through the vasculature of plants from the shoot to the root (11). Interestingly, in recent years it has been shown that sRNAs can also be transmissible between organisms of the same species but predominantly between hosts and pathogens, parasites or symbionts. In most cases these sRNAs regulate genes that determine susceptibility or resistance. For example, the human pathogen Trypanosoma cruzi produces extracellular vesicles containing tRNA-derived sRNAs that regulate susceptibility to infection of mammalian cells (12). On the other hand, miRNAs from human erythrocytes are responsible for resistance against the malaria parasite Plasmodium falciparum (13).
Plant pathogens seem to be very versatile and use similar strategies in order to facilitate their infection. Molecular pathogens like viruses or viroids use the RNA silencing pathway that was originally meant for protection against invading RNAs to produce host gene-targeting sRNAs. For example, the Cucumber mosaic virus (CMV) produces an sRNA that targets the chlorophyll biosynthetic gene CHLI leading to the production of its characteristic yellowing symptoms (14,15). Botrytis cinerea, the fungus that causes the gray mold disease, produces sRNAs that are loaded into the host AGO1 and silence genes that increase the susceptibility to its infection (16). Defense against this same fungal agent by the host involves the inclusion of plant sRNAs regulating fungal pathogenicity into extracellular vesicles that are transported into the fungal pathogen (17).
MicroRNAs in parasitic dodder – the ammunition in the battle between host and parasite?
“Dodders” is the common name for a group of parasitic plants of the angiosperm genus Cuscuta that looks rather unlike a normal plant (Figure 1A). Having been reduced to what is necessary for its survival and propagation, it has only stems to wrap around their hosts, feeding organs known as “haustoria” to secure the supply with nutrients, and flowers for reproduction. Photosynthetic carbon fixation is negligible (18). Every infection starts with the parasite coiling around the host stem (Figure 1B) and gluing itself onto its surface with a sticky mucilage. The developing haustorium then breaks through the host plant surface and grows until it has reached the vascular tissue. Mature Cuscuta haustoria produce lateral and apical finger-like scavenging cells called “feeding hyphae” that meander between the host cells. Their function is to re-direct water, minerals and organic compounds to the parasite. An intriguing feature of these hyphae is that they differentiate into the cell type they connect with (19), allowing an apparently seamless connection between the two partners. During this process, the Cuscuta haustorium faces a number of challenges. It has to first of all soften or decompose the middle lamellae and possibly primary cell walls of the host, in order to reduce the tissue cohesion and allow its own growth without, however, compromising its own cell wall functions. Enzymes involved in this process include cellulases and xyloglucan endotransglucosylase/hydroxylase (XET) acting on cell wall polymers (20,21). Alas, breakdown products of the cell wall and other damage-associated molecular patterns are known to signal wounding or a pathogen attack to the plant and normally induce defense reactions. For a successful infection, Cuscuta has to inhibit such signaling and take control over the defense response system of its hosts.
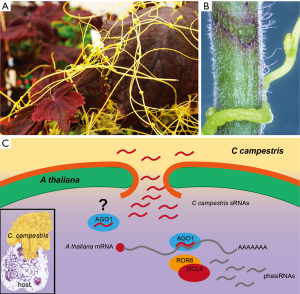
A clue to how the parasite might achieve this was recently provided by Shahid et al. (1). They reasoned that if siRNAs can be easily introduced into the parasite in a process that became known as host-induced gene silencing (HIGS), the opposite might be possible, too. Indeed, when they compared non-infecting Cuscuta campestris (field dodder) stems and uninfected Arabidopsis thaliana stems with the interface consisting of the parasite haustorium inserted into the host tissue, they found 43 upregulated miRNAs at the interface. Forty-two of these were novel miRNAs that lack sufficient sequence similarity to align with any known miRNAs (1). Their further studies suggest that C. campestris deploys these miRNAs into A. thaliana during the infection (Figure 1C). A majority of these miRNAs are from a special class of 22 nts (instead of the regular size for plants of 21 nts), which are known to induce the production of phasiRNAs from their target mRNAs. Although C. campestris seems to take advantage of this amplification effect by hijacking the host’s secondary siRNA production machinery (Figure 1C), the direct effect of the own deployed miRNAs appears to be most effective (1).
Six mRNAs from A. thaliana were identified as targets of the trans-species gene silencing. Three of them encode auxin receptors and one encodes a membrane-localized kinase. For these four, roles in pathogen-induced signaling and plant development are implicated, indicating that these receptors and kinases could be part of the Cuscuta-detection machinery of the hosts. Another target mRNA encodes a predicted transcriptional repressor. The sixth target mRNA, finally, codes for a photosynthate sequestration regulator. Its downregulation, fittingly, could help the parasite to redirect nutrients to its own growing tissue.
Coincidence or evolutionary advantage?
C. campestris has been observed to infect a large range of hosts from many families across the angiosperm spectrum, including herbaceous plants, shrubs and trees (22). Moreover, multiple host species can be infected simultaneously by one and the same individual. This lack of host specificity entails that Cuscuta is likely to use mechanisms to break through the host tissue, silence defence mechanisms and redirect nutrients that are effective in many, if not all, hosts.
Shahid et al. (1) have tested whether the miRNAs upregulated in the haustoria of C. campestris are also effective in silencing of gene expression in other species than in A. thaliana. They found that four of the mRNA target sequences are conserved in multiple species, indicating that the corresponding miRNAs could act on a range of potential hosts. Indeed, Nicotiana benthamiana, when infected with C. campestris, showed a similar reduced accumulation of TIR1 and BIK1 RNAs, which are predicted targets of the released parasitic miRNAs, as A. thaliana.
In summary, this work nicely reveals that the parasitic plant Cuscuta has evolved miRNAs to act on its angiosperm hosts in a trans-species manner and manipulate their defence responses and/or the flow of nutrients. It will be interesting to see whether the same miRNAs are used also by other Cuscuta species and if differences in these miRNAs relate to differences in the host spectra. If the identified miRNAs are indeed solely produced with the intent to manipulate the host and lack primary targets in C. campestris itself, as was suggested by Shahid et al. (1), the question is whether they evolved from vertically inherited genes or were horizontally acquired from one of their hosts. Also, it will be interesting to see from which time point on the miRNAs are exchanged and active since the transport pathway is not yet understood. If miRNAs are exchanged through phloem connections as it seems to be the current understanding, they would not be able to act in their hosts before the connection is already well established and would thus not reach their full potential. At the same time it is unknown if there is a reciprocal transport of miRNAs from the host into C. campestris that could also be biologically active. With the recent publication of the genome sequence of C. campestris (22), followed closely by that of Cuscuta australis (23), these and other questions will without doubt be answered soon.
Acknowledgments
Funding: Work on Cuscuta-host interactions in the lab of K. Krause is supported by the Tromsø Research Foundation. Work on RNA silencing in the lab of G. Martinez is supported by the Swedish Research Council, the Swedish University of Agricultural Sciences and the Carl Tryggers Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Huanyu Gu (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.07.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shahid S, Kim G, Johnson NR, et al. MicroRNAs from the parasitic plant
Cuscuta campestris target host messenger RNAs. Nature 2018;553:82-5. [Crossref] [PubMed] - Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 2015;16:727-41. [Crossref] [PubMed]
- Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 2011;12:221. [Crossref] [PubMed]
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 2013;11:745-60. [Crossref] [PubMed]
- Yu Y, Jia T, Chen X. The 'how' and 'where' of plant microRNAs. New Phytol 2017;216:1002-17. [Crossref] [PubMed]
- Martinez G, Köhler C. Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr Opin Plant Biol 2017;36:22-8. [Crossref] [PubMed]
- D'Ario M, Griffiths-Jones S, Kim M. Small RNAs: Big Impact on Plant Development. Trends Plant Sci 2017;22:1056-68. [Crossref] [PubMed]
- Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013;25:2400-15. [Crossref] [PubMed]
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in
Caenorhabditis elegans . Nature 1998;391:806-11. [Crossref] [PubMed] - Voinnet O, Vain P, Angell S, et al. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 1998;95:177-87. [Crossref] [PubMed]
- Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J 2011;30:3553-63. [Crossref] [PubMed]
- Garcia-Silva MR. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol Res 2014;113:285-304. [Crossref] [PubMed]
- LaMonte G, Philip N, Reardon J, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012;12:187-99. [Crossref] [PubMed]
- Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 2011;7:e1002022 [Crossref] [PubMed]
- Shimura H, Pantaleo V, Ishihara T, et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 2011;7:e1002021 [Crossref] [PubMed]
- Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013;342:118-23. [Crossref] [PubMed]
- Cai Q, Qiao L, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018;360:1126-9. [Crossref] [PubMed]
- van der Kooij TA, Krause K, Dörr I, et al. Molecular, functional and ultrastructural characterisation of plastids from six species of the parasitic flowering plant genus
Cuscuta . Planta 2000;210:701-7. [Crossref] [PubMed] - Christensen NM, Dörr I, Hansen M, et al. Development of Cuscuta species on a partially incompatible host: induction of xylem transfer cells. Protoplasma 2003;220:131-42. [Crossref] [PubMed]
- Johnsen HR, Krause K. Cellulase activity screening using pure carboxymethylcellulose: application to soluble cellulolytic samples and to plant tissue prints. Int J Mol Sci 2014;15:830-8. [Crossref] [PubMed]
- Olsen S, Popper ZA, Krause K. Two sides of the same coin: Xyloglucan endotransglucosylases/hydrolases in host infection by the parasitic plant
Cuscuta . Plant Signal Behav 2016;11:e1145336 [Crossref] [PubMed] - Vogel A, Schwacke R, Denton AK, et al. Footprints of parasitism in the genome of the parasitic flowering plant
Cuscuta campestris . Nat Commun 2018;9:2515. [Crossref] [PubMed] - Sun G, Xu Y, Liu H, et al. Large-scale losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat Commun 2018;9:2683. [Crossref] [PubMed]
Cite this article as: Martinez G, Krause K. The parasitic plant haustorium: a trojan horse releasing microRNAs that take control of the defense responses of the host. Non-coding RNA Investig 2018;2:44.


