
Unveiling the protein coding-independent function of the TET family in gastric cancer
The expression of a specific gene is epigenetically regulated at multiple steps at the transcriptional and post-transcriptional levels under physiological conditions. At the transcriptional level, an “open” or “closed” chromatin structure of the promoter region of a gene is an important factor for inducing or repressing transcription (1). Epigenetic hallmarks such as DNA methylation and histone modification (for example, methylation, acetylation, and ubiquitination) levels are closely related to the chromatin environment (2). In pathological conditions such as cancer, epigenetic hallmarks at the promoters of tumor-suppressor genes or oncogenes are dysregulated, leading to abnormal gene expression patterns.
DNA methylation, in particular methylation at cytosine residues (5-Methylcytosine, 5 mC) in CpG (5'-C-phosphate-G-3') sequences, is essential for epigenetic regulation and for maintaining genomic stability (2). CpG methylation induced by DNA methyltransferases (DNMTs) leads to the recruitment of various methyl-CpG-binding proteins to alter the chromatin structure and therefore repress transcription (2). DNA methylation patterns across the human genome are dramatically changed in various disorders. We have previously reported that infection with Helicobacter pylori, which is thought to be involved in gastric cancer risk, induces aberrant DNA methylation in gastric epithelial cells, contributing to genome-wide gene expression changes (3).
Regarding DNA demethylation, ten-eleven-translocation-1 (TET-1) was identified as a DNA demethylase in mammals (4), and shown to catalyze the successive oxidation of 5 mC to 5-hmC (5-hydroxymethylcytosine), 5-fC (5-formylcytosine), and 5-caC (5-carboxylcytosine). These oxidative products could be excised by TDG (thymine DNA glycosylase) and repaired to produce demethylated cytosine. Several members of the TET family of proteins (TET-1, -2, and -3) play important roles in carcinogenesis and tumor progression in a variety of human malignancies (5). Global decreases in TET protein expression cause low levels of 5-hmC and thereby enhance promoter hypermethylation and promote tumorigenesis; these results imply that TET proteins are tumor suppressors (6,7). However, TET1 is overexpressed in MLL-rearranged leukemia and plays a critical oncogenic role (8). The TET family of proteins has diverse and complex biological effects at the transcriptional level, and serve as tumor-suppressor genes or oncogenes in a context-dependent manner.
After transcription, mRNA (messenger RNA) is regulated at the post-transcriptional level until the initiation of translation and during translation. The 3'-untranslated region (3'UTR) located after the stop codon of mRNA contains many elements for gene regulation (9). microRNA (miRNA), an endogenous small non-coding RNA molecule, represents a typical example of 3'UTR-mediated gene regulation. It is currently well known that miRNAs complementarily bind to the 3'UTR of target mRNA and repress its expression at the post-transcriptional level. This process occurs in development, differentiation, and diseases. While mRNA is actively being transcribed, protein expression could be strongly repressed by miRNA, leading to a discrepancy of a high mRNA level but low protein level (10). Accumulating evidence has shown that changes in miRNA expression dysregulate target gene expression, and is involved in cancer development and progression. For example, we have shown that Brm (SMARCA2), a catalytic subunit of the SWI/SNF chromatin remodeling complex, is actively transcribed but repressed at the post-transcriptional level by miR-199a in several human cancer cell lines (11). This post-transcriptional dysregulation of Brm mRNA by miR-199a is related to anchorage-independent tumor growth (12).
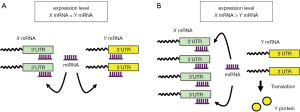
It is estimated that each miRNA has more than 100 target genes; therefore, miRNAs are considered to be global gene regulators. Consequently, miRNAs play a “leading part” in the regulatory system, whereas the target genes are passively repressed. However, a novel concept, “competing endogenous RNA (ceRNA)” has recently emerged, whereby the target genes regulate miRNA activity and therefore play the “leading part” (13). Figure 1A and B shows a simplified model of this concept, showing one miRNA and its target mRNAs, X and Y. If the expression of both X and Y are at the same level, then the miRNA should have equal access to and repress the expression of these two mRNAs (Figure 1A). If the expression of only X is increased for some reason (for example, because of transcriptional activation), the miRNA gains increased access to the abundant X mRNA (Figure 1B). As a result, Y escapes the inhibitory effect of the miRNA and is efficiently translated into Y protein. X is called the ceRNA because it is titrating miRNA away from Y. Some mRNAs, such as PTEN, have already been reported to have this protein-coding-independent function, and play important biological roles in cancer (14).
In this Editorial, we introduce the finding that TET also has a ceRNA function; this finding was recently published in Cancer Research in an article entitled “TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis” (15). The authors demonstrated that several members of the TET family (that is, TET-1, -2, and -3) of mRNAs act as oncogenic ceRNA for the tumor-suppressor miR-26 in gastric cancer. To elucidate the function of TET, they first showed by RT-qPCR that TET mRNA expression is increased in gastric cancer compared with normal tissue (Figure 2). Moreover, high levels of TET family mRNAs were associated with tumor invasion depth, clinical stage, lymph node metastasis, and poor overall survival in gastric cancer. In vitro and vivo experiments, knockdown using lentiviral short-hairpin RNA (shRNA) against each TET family member inhibited gastric cancer cell proliferation and tumor growth, implicating their oncogenic functions. Interestingly, the overexpression of shRNA-resistant TET cDNAs (which lack 3'UTR regions) failed to rescue the inhibition of tumor proliferation induced by the shRNA (which targets the 3'UTRs). This result indicates that the oncogenic function of TET members is independent of their protein-coding potential. In experiments using several TET truncated-cDNA constructs, the 3'UTR region of each TET member was indispensable for their oncogenic role, and the authors hypothesized that the 3'UTRs act as a ceRNA for the miRNA. Moreover, immunohistochemical staining experiments revealed that the protein expression levels of TET family members were almost the same between normal tissues and gastric cancer tissues despite the higher mRNA levels in the tumor tissues, as mentioned above. The discrepancy between mRNA and protein expression levels implies that the TET family of mRNAs are transcribed but might be repressed by miRNAs at the post-transcriptional level.
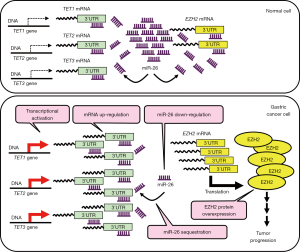
In their previous report, a tumor-suppressive microRNA, miR-26, was shown to be down-regulated in gastric cancer compared with normal tissues, and the target mRNA FGF9 is induced in cancer tissue (16). In the current study, the authors showed that the 3'UTRs of the TET family of mRNAs are directly targeted by miR-26. That is, the 3'UTRs sequestered miR-26 in gastric cancer cells (Figure 2). Overexpression of wild-type TET1/2/3 3'UTRs induced gastric tumorigenesis in vitro and in vivo, whereas mutations in the miR-26 binding sites in TET 3'UTRs abrogated this oncogenic phenotype. Among the previously reported target genes of miR-26 target, they focused on EZH2, which is a potent oncogene in a wide range of human malignancies. Overexpression of TET 3'UTRs sequestered miR-26, and EZH2 was released from miR-26-mediated repression. This effect induced EZH2 overexpression and gastric tumorigenesis (Figure 2).
Given that TET was originally identified as a DNA demethylase, this study is interesting because it reveals the non-coding oncogenic function of TET mRNA in gastric cancer. The authors demonstrated that TET1 mRNA acts as a tumor-promoting ceRNA. Importantly, TET1 protein inhibited gastric cancer growth in their experiments. The opposing roles that TET1 mRNA and TET1 protein play in gastric cancer should help further elucidate the diverse molecular functions. However, it is notable that these results contradict other reports. An analysis of two different cohorts in that study showed that all TET family mRNAs were up-regulated in gastric cancer compared with normal tissue. Conversely, Yang and colleagues showed a decreased expression of TET1 mRNA in gastric cancer (17). Considering that the development and progression of gastric cancer are related to various factors, as summarized previously in our review (18), this contradictory result might stem from differences in clinical and pathological backgrounds, such as a Helicobacter pylori infection. Larger cohort analyses in cooperation with surgeons and pathologists are needed to gain a comprehensive understanding of the expression and the molecular mechanisms of the TET family of mRNAs in gastric cancer.
Through advances in high-throughput sequencing technologies, many novel classes of non-coding RNAs other than miRNAs have been discovered during the last few years; for example, long non-coding RNAs (lncRNAs). lncRNAs are more than 200 nt in length and have no protein-coding potential but exhibit various biological functions (19,20). Some lncRNAs are known to associate with transcriptional and chromatin remodeling factors in the nucleus to recruit them to a specific gene promoter region (19). Meanwhile, other lncRNAs could act as a ceRNA for miRNA in the cytoplasm (21). Yang et al. reported that a lncRNA, SNHG6, which is localized in both the nucleus and the cytoplasm, is overexpressed in gastric cancer cells and induces epithelial-mesenchymal transition (EMT) (22). In addition, a nuclear-localized SNHG6 associates with EZH2 to recruit it to its target promoter region. Meanwhile, Cao and colleagues showed that cytoplasmic-localized SNHG6 sequesters miR-26 to promote hepatocellular carcinoma progression (23). It is interesting to determine whether and how the TET/miR-26/EZH2 axis overlaps with other ceRNA (including coding and non-coding RNA)-related pathways.
Gastric cancer is the fourth most prevalent cancer and the second leading cause of cancer-related death. Considering that many miRNAs are dysregulated in gastric cancer, ceRNA might play an important role in gastric cancer development and progression (24,25). Although further studies are needed to reveal the detail molecular mechanisms, controlling miRNA activity through ceRNA will be a promising therapeutic strategy for gastric cancer and other malignancies.
Acknowledgments
Funding: This study was supported, in part, by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (16K19090 to K.S. and 15K08960 to T.T.).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.03.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gaspar-Maia A, Alajem A, Meshorer E, et al. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 2011;12:36-47. [Crossref] [PubMed]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 2002;3:662-73. [Crossref] [PubMed]
- Niwa T, Tsukamoto T, Toyoda T, et al. Inflammatory Processes Triggered by Helicobacter pylori Infection Cause Aberrant DNA Methylation in Gastric Epithelial Cells. Cancer Res 2010;70:1430-40. [Crossref] [PubMed]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009;324:930-5. [Crossref] [PubMed]
- Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 2016;30:733-50. [Crossref] [PubMed]
- An J, González-Avalos E, Chawla A, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 2015;6:10071. [Crossref] [PubMed]
- Thienpont B, Steinbacher J, Zhao H, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016;537:63-8. [Crossref] [PubMed]
- Huang H, Jiang X, Li Z, et al. TET1 plays an essential oncogenic role in MLL -rearranged leukemia. Proc Natl Acad Sci USA 2013;110:11994-9. [Crossref] [PubMed]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5' - and 3' -UTR-binding factors. Trends Biochem Sci 2003;28:182-8. [Crossref] [PubMed]
- Loayza-Puch F, Yoshida Y, Matsuzaki T, et al. Hypoxia and RAS-signaling pathways converge on, and cooperatively downregulate, the RECK tumor-suppressor protein through microRNAs. Oncogene 2010;29:2638-48. [Crossref] [PubMed]
- Sakurai K, Furukawa C, Haraguchi T, et al. MicroRNAs miR-199a-5p and -3p Target the Brm Subunit of SWI / SNF to Generate a Double-Negative Feedback Loop in a Variety of Human Cancers. Cancer Res 2011;71:1680-9. [Crossref] [PubMed]
- Kobayashi K, Sakurai K, Hiramatsu H, et al. The miR-199a/Brm/EGR1 axis is a determinant of anchorage-independent growth in epithelial tumor cell lines. Sci Rep 2015;5:8428. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA Hypothesis : The Rosetta Stone of a Hidden RNA Language ? Cell 2011;146:353-8. [Crossref] [PubMed]
- Karreth FA, Tay Y, Perna D, et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell 2011;147:382-95. [Crossref] [PubMed]
- Deng M, Zhang R, He Z, et al. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer Res 2017;77:6069-82. [Crossref] [PubMed]
- Deng M, Tang H, Lu X, et al. miR-26a Suppresses Tumor Growth and Metastasis by Targeting FGF9 in Gastric Cancer. PLoS One 2013;8:e72662 [Crossref] [PubMed]
- Yang Q, Wu K, Ji M, et al. Decreased 5-hydroxymethylcytosine (5-hmC) is an Independent Poor Prognostic Factor in Gastric Cancer Patients Independent Poor Prognostic Factor in Gastric Cancer Patients. J Biomed Nanotechnol 2013;9:1607-16. [Crossref] [PubMed]
- Tsukamoto T, Nakagawa M, Kiriyama Y, et al. Prevention of Gastric Cancer : Eradication of Helicobacter pylori and Beyond. Int J Mol Sci 2017;18. [PubMed]
- Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013;14:699-712. [Crossref] [PubMed]
- Sakurai K, Reon BJ, Anaya J, et al. The lncRNA DRAIC / PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res 2015;13:828-38. [Crossref] [PubMed]
- Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics 2016;14:73-80. [Crossref] [PubMed]
- Yan K, Tian J, Shi W, et al. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem 2017;42:999-1012. [Crossref] [PubMed]
- Cao C, Zhang T, Zhang D, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017;36:1112-22. [Crossref] [PubMed]
- Cao D, Jia Z, You L, et al. 18β-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget 2016;7:71960-73. [Crossref] [PubMed]
- Hao NB, He YF, Li XQ, et al. The role of miRNA and lncRNA in gastric cancer. Oncotarget 2017;8:81572-82. [Crossref] [PubMed]
Cite this article as: Sakurai K, Tsukamoto T. Unveiling the protein coding-independent function of the TET family in gastric cancer. Non-coding RNA Investig 2018;2:17.


