
lncRNA-ACOD1: GOT2 has got a lncRNA cofactor
In spite of the limited information encoded in viral genomes, viruses can infect and replicate in complex organisms. Evolution has selected viruses able to use cellular factors to achieve viral entry, viral protein expression, and viral replication, packaging and release. In addition, during viral cell-cycle, viruses need to inhibit the cellular antiviral response. All these processes require a considerable amount of energy and resources, which need to be stolen from cellular machineries. Therefore, viruses also need to manipulate the metabolism of the host cell (1).
Interactions between host cells and viruses involve protein factors, micro RNAs (miRNAs) and long noncoding RNAs (lncRNAs). LncRNAs are transcripts longer than 200 nts with poor coding capacity and good potential to function as RNA molecules. In the last decade, several groups have shown lncRNA genes to be quite numerous in mammalian genomes (2,3). LncRNAs can accumulate in the nucleus or in the cytoplasm of the cell. Several nuclear lncRNAs have been shown to play regulatory functions by modulating chromatin remodeling, promoter-enhancer interactions, transcription or splicing (4). In the cytoplasm, some lncRNAs have been described to regulate mRNA translation and stability, or serve as miRNA sponges decreasing miRNA availability and functionality (5). The function of some of the few lncRNAs studied up to date is required for essential cellular processes such as cell proliferation, differentiation, signaling or return to homeostasis (5,6). In addition, several lncRNAs have been described to exert their regulatory properties on virus infected cells (7).
The transcriptome of the infected cell shows altered levels of many lncRNAs (7-10). Adenovirus, Epstein-Barr virus, Herpes or Flavivirus express noncoding transcripts required for efficient viral replication. Therefore, some lncRNAs that accumulate in the infected cell are from viral origin. Others are cellular lncRNAs that may be induced or repressed after infection. Expression of these cellular lncRNAs may be altered by viral factors or by the cellular response to viral infection. In fact, several lncRNAs have been described as induced by the pathways activated in the cell after detection of pathogen-associated molecular patterns. These pathways include activation of the transcription factors IRF3 and NF-κB, and the type I interferon (IFN) response. Interestingly, some lncRNAs induced by the antiviral/IFN response function to regulate the antiviral pathway. Examples are lncRNA BISPR, which is a positive regulator, or NRIR or EGOT, which are negative regulators of the IFN response that may allow the cell return to homeostasis (9,11,12). Given so, lncRNA BISPR exerts antiviral properties while NRIR or EGOT help viral infection by inhibiting the antiviral pathway. Recently, a novel lncRNA-mediated mechanism to promote viral infection has been published in Science by the team of Dr. Xuetao Cao (13).
Wang and colleagues analyzed the transcriptome of controls, wild-type or IFN-I receptor (IFNAR)-deficient macrophages infected with Vesicular Stomatitis Virus (VSV) and identified a collection of viral-induced lncRNAs that are not regulated by IFN. One of them is lncRNA-ACOD1, an ACOD1 neighboring gene. LncRNA-ACOD1 is induced by VSV, as well as by other RNA and DNA viruses such as Sendai virus (SeV), Herpes simplex virus (HSV-1) or Vaccinia virus (VACV) in different cell lines (Figure 1). As mentioned above, lncRNA-ACOD1 is not induced by IFN-I or by the IRF3 transcription factor. Instead, sensing of the virus activates the NF-κB pathway whose p65/RelA subunit can bind the promoter of lncRNA-ACOD1 and induce its transcription (Figure 1). Then, induction of NF-κB will on one hand alert the immune system about the infection inducing the expression of proinflammatory factors, while on the other hand, it will upregulate lncRNA-ACOD1 which, paradoxically enough, functions as a proviral factor.
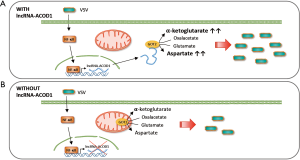
Knock down of lncRNA-ACOD1 convincingly decreases VSV, HSV-1 and VACV viral load by affecting viral replication. These results were obtained with cultured macrophages after lncRNA-ACOD1 decrease with RNAi or gene editing with the CRISPR-Cas9 system, or in lncRNA-ACOD1-/- mice. Indeed, lncRNA-ACOD1 knocked out animals survived after infection with a lethal dose of VSV while wild-type controls die. LncRNA-ACOD1 depletion does not block viral entry, does not increase the IRF3-IFN-I antiviral response, and does not influence cell apoptosis or autophagy, which are cellular mechanisms that affect productive viral infections. Further, while some nuclear lncRNAs function in cis to regulate the expression of neighboring genes (14), lncRNA-ACOD1 does not. Inhibition of lncRNA-ACOD1 by RNAi did not affect the expression levels of the closest genes Kctd12 and Acod1. In addition, inhibition of Kctd12 or Acod1 did not affect lncRNA-ACOD1 expression or VSV levels after infection, suggesting that lncRNA-ACOD1 effect upon VSV infection is not mediated by regulating nearby genes. In fact, lncRNA-ACOD1 is spliced, polyadenylated and accumulates preferentially in the cytoplasm.
The authors obtained the first clues about lncRNA-ACOD1 function after transcriptome analysis of lncRNA-ACOD1 depleted and non-depleted cells. The loss of lncRNA-ACOD1 caused a significant deregulation of many metabolism-related genes. Metabolomic analysis by LC-MS/MS showed that the metabolic changes observed after infection of wild-type cells (1), were not detected in lncRNA-ACOD1-depleted cells. Although lncRNA-ACOD1 is not highly expressed (~100 copies per cell), the endogenous lncRNA complexes were purified following a modified chromatin isolation by RNA purification, and mass spectrometry was used to determine that lncRNA-ACOD1 is bound by glutamic-oxaloacetic transaminase 2 (GOT2) (Figure 1). LncRNA-ACOD1-GOT2 binding was confirmed by RNA immunoprecipitation with GOT2 specific antibodies.
GOT is a pyridoxal phosphate-dependent aminotransferase enzyme that plays a role in amino acid metabolism, in the urea and tricarboxylic acid cycles and facilitates cellular uptake of long-chain free fatty acids (15). GOT catalyzes the conversion of oxaloacetate and L-glutamate into aspartate and alpha-ketoglutarate, and therefore it is involved in the mitochondrial malate-aspartate shuttle (16). Two homodimeric enzymes exist, GOT1 and GOT2, which show close homology. While GOT1 localizes to the cytoplasm, GOT2 accumulates in the inner mitochondrial membrane (17). Surprisingly, the interaction between lncRNA-ACOD1 and GOT2 occurs in the cytoplasm (Figure 1). It seems currently unknown whether lncRNA-ACOD1 binds fresh GOT2 after translation impeding GOT2 mitochondrial transport. To determine whether binding of lncRNA-ACOD1 and GOT2 plays a relevant role in infection, Wang and colleagues show that: (I) GOT2 knockdown reduces VSV replication; (II) lncRNA-ACOD1 does not affect GOT2 expression; (III) lncRNA-ACOD1 overexpression increases viral replication in wild-type but not in GOT2-depleted cells and; (IV) recombinant GOT2 rescues the defect in virus replication of lncRNA-ACOD1 or GOT2-depleted cells. As a whole, these results indicate that GOT2 mediates the functional effect of lncRNA-ACOD1 on viral replication.
Wang and colleagues made elegant technical efforts to identify lncRNA-ACOD1 interaction domains. RNA sequencing of the protected lncRNA-ACOD1 complex revealed that GOT2 is bound by the 5´-end of lncRNA-ACOD1 (~nts 165–390). MS identification of the protease-digested lncRNA-ACOD1 complex defined a 15 aa peptide (residues 54–68) as the GOT2 region that binds lncRNA-ACOD1. The role of these binding domains was confirmed in experiments with full-length and binding-domain-deleted versions of lncRNA-ACOD1 or GOT2. Interestingly, the region of GOT2 that binds lncRNA-ACOD1 is very close to the substrate binding region, indicating that lncRNA-ACOD1 could affect GOT2 enzymatic activity. Experiments with in vitro synthesized lncRNA-ACOD1 and recombinant GOT2 protein show that this is indeed the case: lncRNA-ACOD1 enhances the catalytic activity of GOT2 (Figure 1). Similar results were observed in vivo. Interestingly, GOT2 metabolites L-aspartate and α-ketoglutarate were reduced in lncRNA-ACOD1 knocked-out mice, and, in these KO animals, injection of L-aspartate and α-ketoglutarate restores an efficient VSV replication.
Although most experiments are performed with mouse cells, Wang et al., show that similar mechanisms could function in human cells. The authors identify a putative human orthologue that does not follow perfect synteny with the mouse counterpart. However, human lncRNA-ACOD1 is induced by influenza virus infection and human lncRNA-ACOD1 depletion by RNAi decreases viral replication. In addition, the region of GOT2 that binds mouse lncRNA-ACOD1 is evolutionary conserved, and the human lncRNA-ACOD1 also binds GOT2.
Taken together, the refined work performed by Wang and colleagues shows that lncRNA-ACOD1 is induced in infected cells to increase GOT2 activity, and provide enough resources to support the viral cell cycle (Figure 1). Increased L-aspartate and α-ketoglutarate will serve to build non-essential aminoacids, nucleotides and energy required for replication. Therefore, this work is a lesson about the metabolic changes induced by viral infection. In addition, Dr. Cao’s team describes a novel function for lncRNAs apart from regulation of gene expression. They show for the first time that lncRNAs can serve as enzyme cofactors. Cofactors are factors required for enzymes to show full activity. Known cofactors are inorganic or organic molecules required to form active holoenzymes. This seminal work describes the first lncRNA cofactor. In fact, it has been known for years that several metabolic enzymes have RNA-binding capacity but it was not clear whether RNA binding could affect functionality (18). More efforts should be now focused on determining whether other lncRNAs function as cofactors of different metabolic enzymes.
Additional experiments will be required to address the role of lncRNA-ACOD1 in non-infected cells. It is difficult to believe that cells express lncRNA-ACOD1 just to help viral infections. Alternatively, lncRNA-ACOD1 could be a switch to increase cellular metabolic resources in specific cellular conditions. Inflammation could be one of them, given that lncRNA-ACOD1 is induced by NF-κB. Other conditions could be the controlled cell proliferation required for immunity or regeneration, or the uncontrolled proliferation that allows tumor formation. It will be interesting to study these processes in the lncRNA-ACOD1-/- mice. These experiments will also help to determine whether lncRNA-ACOD1 could be used as a therapeutic target for the treatment of viral infections and other pathologies.
Acknowledgments
We want to thank Juan Pablo Unfried for helpful comments.
Funding: Spanish Department of Science (SAF2015-70971-R), grant Ortiz de Landazuri from the Government of Navarra and European FEDER funding.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Guoping Li (Cardiovascular Division of the Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.02.01). PF serves as an unpaid editorial board member of Non-coding RNA Investigation from December 2017 to November 2019. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goodwin CM, Xu S, Munger J. Stealing the Keys to the Kitchen: Viral Manipulation of the Host Cell Metabolic Network. Trends Microbiol 2015;23:789-98. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799-816. [Crossref] [PubMed]
- Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 2017. [Epub ahead of print].
- Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 2017;18:962-72. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Fortes P, Morris KV. Long noncoding RNAs in viral infections. Virus Res 2016;212:1-11. [Crossref] [PubMed]
- Carnero E, Fortes P. HCV infection, IFN response and the coding and non-coding host cell genome. Virus Res 2016;212:85-102. [Crossref] [PubMed]
- Carnero E, Barriocanal M, Prior C, et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 2016;17:1013-28. [Crossref] [PubMed]
- Barriocanal M, Fortes P. Long Non-coding RNAs in Hepatitis C Virus-Infected Cells. Front Microbiol 2017;8:1833. [Crossref] [PubMed]
- Barriocanal M, Carnero E, Segura V, et al. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front Immunol 2015;5:655. [Crossref] [PubMed]
- Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 2014;42:10668-80. [Crossref] [PubMed]
- Wang P, Xu J, Wang Y, et al. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017;358:1051-5. [Crossref] [PubMed]
- Yan P, Luo S, Lu JY, et al. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin Genet Dev 2017;46:170-8. [Crossref] [PubMed]
- Inoue K, Kuramitsu S, Okamoto A, et al. Site-directed mutagenesis of Escherichia coli aspartate aminotransferase: role of Tyr70 in the catalytic processes. Biochemistry 1991;30:7796-801. [Crossref] [PubMed]
- Yang H, Zhou L, Shi Q, et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J 2015;34:1110-25. [Crossref] [PubMed]
- Jiang X, Chang H, Zhou Y. Expression, purification and preliminary crystallographic studies of human glutamate oxaloacetate transaminase 1 (GOT1). Protein Expr Purif 2015;113:102-6. [Crossref] [PubMed]
- Castello A, Hentze MW, Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol Metab 2015;26:746-57. [Crossref] [PubMed]
Cite this article as: Barriocanal M, Fortes P. lncRNA-ACOD1: GOT2 has got a lncRNA cofactor. Non-coding RNA Investig 2018;2:9.


