
ciRS-7 acts as a master player in colorectal cancer
Colorectal cancer (CRC) represents a critical health burden worldwide (1,2). Although patients with localized CRC have a 5-year survival rate of about 90%, this ratio dramatically drops in patients with metastatic tumors (2). Standard and liquid biopsy are extremely effective for the identification of critical genetic lesions and to study clonal evolution in CRC (3,4). However, only a minor fraction of oncogenic mutations are clinically actionable, indicating that other novel therapeutic strategies are urgently needed.
The recent discovery that our genome is pervasively transcribed (5) opens a novel scenario in which non-coding RNA (ncRNA) molecules can potentially play a pivotal role in the control of cellular homeostasis and transformation. Circular RNAs (circRNAs) represent a new class of ncRNA whose production is finely regulated during development and differentiation (6). CircRNAs originate from the non-canonical splicing of both coding and ncRNAs in which an upstream splice acceptor is covalently bound to a downstream splice donor in a process called back-splicing (7). While in the last decade, the role of microRNAs (miRNAs) in tumorigenesis has been extensively investigated (8), the impact of other ncRNA species such as circRNAs remains largely unexplored (9,10).
Interestingly, coding and ncRNAs intertwine complex relationships built on base pair complementarity (11). The theory of competing endogenous RNAs (ceRNAs) predicts that any RNA molecule can potentially regulate the level of multiple transcripts by subtracting (or ‘sponging’) miRNAs from other RNAs sharing, throughout their sequence, the same miRNA responsive elements (MREs) (12). CircRNAs, and in particular the circRNA sponge for miR-7 (ciRS-7), epitomize the ceRNA activity (13-15). CiRS-7 originates from the antisense strand of the CDR1 gene and, as any other circRNA, it is particularly stable and resistant to RNA degradation (13,14). However, ciRS-7 acts as a very powerful RNA sponge for miR-7, since its sequence contains more than 70 different MREs for this miRNA (13,14) CiRS-7 is highly abundant in the mammalian brain (13,14). Accordingly, the knock out mouse model develops dysfunction of excitatory synaptic transmission and neuropsychiatric-like alterations (16).
Intriguingly, Weng et al. have recently identified ciRS-7 as a critical circRNA in CRC pathogenesis (17). The analysis of ciRS-7 expression in 153 CRC and 44 matched normal mucosae revealed the over-expression of this circRNA in tumor specimens. The authors further validated this analysis in a cohort of 165 cases. Notably, ciRS-7 was overexpressed in advanced CRC, especially in patients with T4 disease and more advanced stages. Moreover, high ciRS-7 expression correlated with poor overall survival in patients. Since miR-7 is frequently dysregulated in cancer (18) and ciRS-7 acts as a potent miR-7 sponge (Figure 1A), the authors investigated the biological relationship between these two ncRNAs in CRC. In accordance with the ceRNA hypothesis, ectopic expression of miR-7 in CRC cells strongly impaired cell proliferation, migration, invasion and protection from apoptosis. On the other hand, the co-expression of ciRS-7 dramatically reduced the oncosuppressor potential of miR-7 both in vitro and in vivo (xenografts) (Figure 1B). Mechanistically, Weng et al. analyzed two well-established miR-7 target genes, EGFR and RAF1. Both genes were found downregulated at the mRNA and protein level in CRC cells over-expressing miR-7. Conversely, EGFR and RAF1 were not modulated when CRC cells were transfected with miR-7 and ciRS-7 constructs, suggesting that ciRS-7 was acting as a potent sponge for miR-7 (Figure 1A). Finally, the authors analyzed the level of miR-7, ciRS-7, EGFR and RAF1 in CRC specimens. Notably, the levels of expression of ciRS-7 and miR-7 as well as of EGFR/RAF1 and miR-7 were found inversely correlated in CRC specimens, while ciRS-7 expression was found positively associated with EGFR and RAF1 in the same patients.
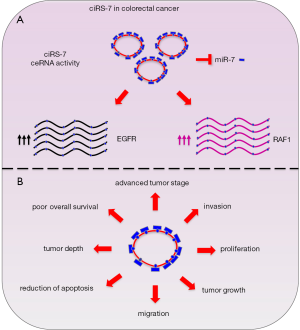
In the last decade the improvement in deep sequencing technologies and bioinformatic analysis has shed light on the biological relevance of the so called “dark matter” of our genome. The work by Weng et al. highlights the impact of circRNAs in cancer pathogenesis. In this specific case ciRS-7 represents either a powerful prognostic biomarker or a future possible therapeutic target for advanced CRC.
The recent discovery that circRNAs can also be efficiently translated (19-21) adds a novel level of complexity, since these peptides can potentially influence gene function at the post-translational level. It is now evident that revealing the complex relationships between coding and ncRNAs in normal and transformed cells represents the crucial step to develop novel therapeutic strategies and diagnostic tools for cancer treatment.
Acknowledgments
We thank Francesca Bersani for helpful discussions and for critically editing the manuscript. Our research is supported by AIRC.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.02.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374-403. [Crossref] [PubMed]
- Siegel RL, Fedewa SA, Anderson WF, et al. Colorectal Cancer Incidence Patterns in the United States, 1974-2013. J Natl Cancer Inst 2017;109: [Crossref] [PubMed]
- Siena S, Sartore-Bianchi A, Garcia-Carbonero R, et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol 2018;29:119-26. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679-92. [Crossref] [PubMed]
- Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res 2016;76:3666-70. [Crossref] [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18. [Crossref] [PubMed]
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555-65. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 2013;20:541-3. [Crossref] [PubMed]
- Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357:eaam8526 [Crossref] [PubMed]
- Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 2017;23:3918-28. [Crossref] [PubMed]
- Gu DN, Huang Q, Tian L. The molecular mechanisms and therapeutic potential of microRNA-7 in cancer. Expert Opin Ther Targets 2015;19:415-26. [Crossref] [PubMed]
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015;21:172-9. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
Cite this article as: Morena D, Taulli R. ciRS-7 acts as a master player in colorectal cancer. Non-coding RNA Investig 2018;2:8.


