The 18 kDa translocator protein, non-coding RNA, and homeostasis
18 kDa mitochondrial translocator protein (TSPO)
In previous studies we found that the 18 kDa mitochondrial TSPO may be at least partly involved in regulation of gene expression of non-coding RNAs (ncRNAs) (1,2). Importantly, modulation of gene expression is not a secondary effect of TSPO activation, but one of its primary functions (3). TSPO is a receptor molecule able to regulate various cellular and organismal functions, in order to maintain health and counteract diseases (4-7). These are functions that can be considered part of the overall homeostatic function of TSPO (4,8). Homeostasis, in this context, stands for any biological system’s activities to maintain a dynamic equilibrium for health and survival (9).
A previous common name for TSPO was peripheral benzodiazepine receptor (PBR), based on its capability to bind benzodiazepines in peripheral tissues (10). Classical TSPO ligands are the benzodiazepine Ro5-4864 (4’-chlorodiazepam) and the isoquinoline carboxamide derivative PK 11195 (11). The primary intracellular location of TSPO typically is the outer mitochondrial membrane (12,13). Numerous reports have demonstrated that TSPO serves various essential functions, including modulation of cell growth and proliferation, ATP production, regulation of the mitochondrial membrane potential (ΔΨm), heme synthesis, mitochondrial cholesterol import associated with steroid synthesis, adaptation to oxidative stress, generation of reactive oxygen species (ROS), modulation of gene expression, and initiation of programmed cell death. The regulatory role of the TSPO in programmed cell death as initiated by ΔΨm collapse is thought to present one mechanism whereby TSPO is involved in cancer and neurodegeneration (14). On occasion, apart from the mitochondria, TSPO can also be found in association with other cell organelles such as the cell nucleus and even the outer cell membrane (4,15,16). The association of the TSPO with the mitochondria-to-cell nucleus signaling pathway is thought to take part in regulation of cell nuclear gene expression (1-3). TSPO knockdown with siRNA as well as application of TSPO ligands indicated that TSPO is important for regulation of gene expression (1-3). In addition, pathway analysis suggested that this gene expression regulation serves various functions including programmed cell death, inflammation, immune response, cell migration, adhesion, cell differentiation, neurite outgrowth, proliferation, tumorigenesis, cell cycle, etc. (3). It can be suggested that modulation of cell nuclear gene expression and regulation of initiation of the mitochondrial apoptosis cascade can be considered main functions of TSPO and its ligands (3,17). This information gave further insights how TSPO could perform its homeostatic functions. In this context, TSPO upregulation has been connected to several diseases, including cancer, neuronal damage, neurodegeneration, and inflammation (16,18,19). Moreover, TSPO ligands can serve to counteract such diseases (7,20).
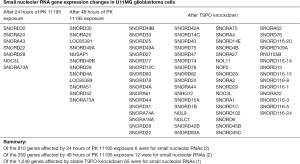
Generation of transgenic mice with TSPO knockout also suggested homeostatic functions of TSPO (21). For example, two distinct types of TSPO knockout mice to induce gonadal and steroidogenic cell-specific TSPO deletion indicated a role for TSPO in cellular cholesterol and lipid homeostasis (22). TSPO expression also appears to be essential for healthy adipocyte functions. In particular, activation of TSPO in adipocytes improves regulation of glucose homeostasis (23). While discussed for mammals, the role of TSPO in homeostasis is well recognized in prokaryotes. In this context it should be noted that TSPO is a protein whose typical trans-membrane helix structure as well as functions have been remarkable preserved along the evolutionary record from Archaea and Bacteria to Eukaryotes (24,25). Studies on prokaryotes suggest that TSPO might be involved in iron homeostasis, synthesis of steroids that regulate membrane fluidity due to its localization, as well as to drive responses to stressors (26). Interestingly, a direct role of TSPO in expression of ncRNAs has been suggested (1,2) (Figure 1). In particular, TSPO appeared to be involved in modulation of nuclear gene expression including small nucleolar (sno) RNAs. Of the 610 genes affected by 24 hours of the TSPO classical ligand PK 11195 exposure, 6 were for small nucleolar RNAs. Of the 359 genes affected by 48 hours of PK 11195 exposure, 12 were for small nucleolar RNAs. Of the 1,249 genes affected by stable TSPO knockdown 65 were for small nucleolar RNAs (1,2).
ncRNAs
ncRNAs are more and more considered as participants in maintaining homeostasis of the host (27,28). While ncRNAs are not translated to proteins, they can modulate protein synthesis in the cell via various post transcriptional modulations, for example, translational repression. The number of ncRNAs within the human genome is known to be higher than RNA coding for proteins. Important functional ncRNAs include: small RNA such as microRNA (miRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), short interfering RNA (siRNA), PIWI-interacting RNA (piRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), extracellular RNA (exRNA), small Cajal body-specific RNA (scaRNA), and long ncRNA (lncRNA). Functions of all of these ncRNAs are summarized briefly below.
miRNAs, which are the most examined members of this family of snRNAs, are small non coding molecules synthesized from the double stranded region of hairpin RNA (29). They are associated with regulation of gene expression via mRNAs translation interference by activating the RNA induced silencing complex (RISC) (30). It has been proven that changes in the level of expression of different types of miRNAs influence the expression of genes responsible for a number of disorders and diseases, including cancer, non-alcoholic fatty liver disease (NAFLD), cardiovascular disease, etc. (31-33).
As TSPO, specific miRNAs appear to play important roles in neurodegenerative diseases and brain injury. Microarray analyses have demonstrated that during mammalian brain development the expression levels of many miRNAs are dynamically controlled, suggesting a role in regulating brain structure and function (34). These same miRNAs can be differentially expressed in the brains of people suffering from CNS-related disorders. For example, various studies have shown significantly increases of miR-9 and miR-128, and a decrease of miR-107 in brains of Alzheimer Disease (AD) patients (35,36). In another study it was found that 15 miRNAs were dysregulated in sporadic AD, in particular increases of miR-197 and miR-511; and decreases of: miR-7i, miR-9, miR-15a, miR-19b, miR-22, miR-26b, miR-29b-1, miR-93, miR-101, miR-106b, miR-181c, miR-210, and miR-363 were found (37). Furthermore, already early in the pathogenesis of AD, miRNA levels can be altered, suggesting that miRNAs take part in the development of the disease (38). Regarding Huntington Disease (HD), miRNAs were found to be differentially expressed in post-mortem HD brains, for example decreases of miR-9 mir-29a, mir-124a, mir-132, and mir-135b expression (39-41). Previous studies have shown that regulation of several miRNAs, including miR-9, miR-29a, and miR132/212, may be directly controlled through proximal binding sites for the REST protein (RE1-silencing transcription factor), potentially presenting a factor for aberrant transcriptional and post-transcriptional control in HD patients (39-42). Regarding Parkinson Disease (PD), from a panel of 224 miRNAs precursors from samples of midbrain, cerebellum, and cerebral cortex samples of PD patients and normal controls, 8 appeared to be enriched in midbrain relative to cerebral cortex or cerebellum, as determined with rtPCR (42). Expression of one of these eight precursor miRNAs, miR-133b was specifically deficient in the context of PD patient samples (42). Numerous studies have shown that brain injury affects expression of coding and ncRNAs, for example, the chronic condition of traumatic brain injury (TBI) can be accompanied by changes in gene expression of over 7,000 genes (involved in metabolism, receptor-mediated cell signaling, neuronal plasticity, immune cell recruitment, immune cell infiltration, and neurodegenerative disease) in the areas of secondary brain injury surrounding the primary injury (43). It can be suggested that miRNAs can be used as biomarkers for these various conditions and diseases.
Regarding the well-known tRNAs and rRNAs, apart from their canonical translational functions, tRNAs can also regulate gene expression (44). Furthermore, amino-acylated tRNAs have been implicated as substrates for non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. tRNA fragments (tRFs) have been recognized to play regulatory roles in translation and gene expression (45,46). Thus far, rRNA is only known as the RNA component of the ribosome (47).
siRNAs are small duplexes that consist of 19–23 paired nucleotides synthesized from double stranded DNA precursors (48,49). These molecules provide similar interference mechanism for silencing a specific gene via RISC as miRNAs. It is reported that siRNAs silencing effect is accomplished via the cooperation with members of the conserved Argonaute (AGO) protein family which enable targeting of complementary RNAs for degradation, translational repression, and transcriptional sequencing (50,51). siRNAs may be used for gene therapy (52).
Compared to miRNAs and siRNAs, piRNAs are somewhat longer molecules consisting of 24–31 nucleotides. They are derived from single stranded precursors (53). The function of piRNAs is enabled via cooperation with specific proteins known as PIWI, for example as expressed in gonads that produce RISCs with a number of RNAs (54). It has been shown that this interaction between PIWI proteins and piRNAs plays a key role in silencing transposable elements (TEs), whose mobility otherwise threatens genome integrity (55).
snRNAs are present in the nucleoplasm and the nucleolus of eukaryote cells. Research by Will and Luhrmann (56) has indicated that snRNAs are involved in the process of splicing of mRNA as elements of the spliceosome making them key players in removing introns and mRNA maturation (56).
As mentioned, we have found in two separate studies that apart from protein coding genes, expression of numerous genes for snoRNAs are affected by TSPO knockdown, and also by the TSPO ligand PK 11195 (1,2). These snoRNA distilled from more than thousand genes affected by TSPO and its ligands are compiled in Figure 1.
snoRNAs derive their name from their location in the nucleolus. snoRNAs primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs (57,58). These post-transcriptional modifications are important for the production of efficient and accurate ribosomes (59). However, snoRNAs can also participate in modifications of snRNAs that mediate mRNA splicing (60). Similar to the splicing snRNAs, snoRNAs are complexed with specific proteins and exist as discrete ribonucleoprotein particles (snoRNPs) (61,62). ScaRNAs are snoRNAs that are localized in Cajal Bodies (CB) which guide the 2-O methylation and pseudouridylation of several snRNAs (63,64). ScaRNAs level of expression is crucial for regular mRNA splicing. Furthermore, snoRNA transcripts serve as the precursors of miRNA-like small RNAs and as regulators of alternative splicing (65,66).
In vitro studies showed that snoRNAs can act either as oncogenes or as tumor suppressors (67). Apart from cancer, snoRNAs may also play a role in neurodegeneration. For example, Snord 3A, a molecular marker and modulator of prion disease progression, was found to be elevated several times in Creutzfeldt Jacobson Disease patients as well as in animal models for the disease (68). Interestingly, SNORDA3 was also found to be affected by TSPO knockdown (Figure 1).
Thus, snoRNAs can participate at several stages of protein formation, including: (I) folding of pre-rRNA as RNA chaperones; (II) protein folding; (III) formation of rRNP substrates; (IV) RNA processing and base modification reactions; (V) assembly of ribosomal subunits; and (VI) export of assembled subunits. Hence it can be considered that optimal expression of these molecules is directly responsible for regular protein synthesis.
In addition to the aforementioned small ncRNAs which are highly abundant molecules, long non coding (lnc)RNA are somewhat larger, having more than 200 nucleotides (69,70). lncRNAs act as blockers of transcriptional and splicing factors, and as modifiers of chromatin protein, thereby regulating the processes of transcription and splicing. It is reported that some antisense lncRNAs can also participate in mRNA stabilization by preventing miRNA binding which makes them potential regulators of their functions as gene silencers (71). In addition, it is also suggested that some lncRNAs regulate Dicer endonuclease activity therefore interfering with siRNA expression (72). Furthermore, lncRNAs have a number of important roles in several biological processes such as, translation, apoptosis, pluripotency of stem cells, etc. (73-76). Studies have also reported the significance of the lncRNA MALAT 1 (full name: ‘metastasis associated lung adenocarcinoma transcript 1’) in reducing inflammation in endothelial cells in diabetic rats (77). Moreover, MALAT 1 silencing also contributed in reducing glucose induced upregulation of inflammatory cytokines such as IL 6 and TNFα (78).
With studies on ncRNA such as discussed above it is becoming well understood that ncRNAs can dramatically affect the execution of myriad cellular programs (79). This study also states that ncRNAs present a complexity as diverse as mRNAs, including a diverse and interrelated range of regulatory and functional roles in transcriptional, post-transcriptional, epigenetic, and nuclear processes. This adds to the cell type-, developmental stage-, and stimulus-specific profiles of protein-coding mRNA expression, post-transcriptional modifications, localization, and translation. Thus, it can be safely concluded that ncRNAs present an intricate frame work that can well serve to maintain homeostasis of biological organisms.
ncRNA, TSPO, and homeostasis
Biological systems are characterized by active maintenance of their homeostasis under broad ranges of environmental and physiological conditions (9). At molecular biological levels, this includes responses to insulting conditions such as injuries, pathogenic infections, and diseases (80-82). Such responses involve coordinated modulation of gene expression programs (83). In this context of homeostasis, long and short ncRNAs regulate development and cell physiology; and are also involved in several human diseases, such as cancer, neurodegenerative disorders; and in a variety of stress responses, including host pathogen interactions (9). Here we want to pay a bit more attention to potential commonalities between TSPO and ncRNA in regulating homeostatic functions.
Various miRNAs play important roles in regulating stress by optimizing the levels of key proteins involved in stress response (84). Several miRNAs, for example miR-9, miR-146, and miR-155 present increased expression in acute stress, acting via negative feedback to alleviate inflammatory reactions (85). Other studies have shown miR-21 to be overexpressed in glioblastoma tumors, described as anti-apoptotic factor predicted to down regulate genes associated with advanced apoptosis (86). In the section of miRNAs it was discussed that miRNAs are participants in mechanisms related to brain injury and brain disease.
The use of aberrant miRNAs offers opportunities to screen, diagnose, and predict patients’ outcome with high accuracy by simple blood testing for different types of cancer (87). In this context, miRNAs have been linked to the regulation of differentiation, proliferation, apoptosis, and exocytosis. These are functions that are also known to be under the control of TSPO, as discussed at the beginning of this review.
Here we want to pay attention to some more functions that imply regulation of homeostasis by TSPO as well as miRNAs. Since TSPO is abundantly expressed in various metabolically active tissues, interest has been directed toward its regulatory role in several important metabolic disorders. Because one of its endogenous ligands is cholesterol, TSPO expression was measured in high fat/high cholesterol fed mice that present an inverse correlation between TSPO expression and obesity related inflammation (88,89). Similarly, a significant decrease in mitochondrial TSPO expression was observed in obese individuals (90). In the same study, in vitro incubation of trophoblast cells with long chain saturated fatty acids, showed inhibited TSPO mRNA expression. Taking this into account, special interest has been shown in TSPO’s role in atherosclerosis and fatty liver disease introducing it as a biomarker for oxidative stress (91). In addition, several studies have revealed that TSPO is highly expressed in white and brown adipose tissue (WAT and BAT), thus implying TSPO’s involvement in adipocytes’ metabolism making TSPO an important factor considering regulation of obesity (92). lncRNAs involvement in cholesterol and triglyceride homeostasis was also shown (93,94). miRNA relation to inflammation as a result of obesity was also observed. For example, Ortega et al. (95) reported significant increases in expression of miR-221, miR-222, and miR-155 under conditions of obesity and their consequent decrease in post weight loss patients (95). Also an association between miR-155 and inflammation has been made by Karkeni et al. (96) who affirmed up-regulation of this miR-155 by TNFα in adipose tissue (96). Jiang et al. (97) reported significant increase of miR-378 in mature adipocytes as a result of IL-6, TNFα, and leptin treatment thus confirming miR-378’s connection to obesity related inflammation (97). Further research exploring the outcome of therapeutic treatment associated with different miRNA profiles could provide valuable data to advance drug regime selection.
It has been postulated that TSPO is involved in cardiovascular diseases as well as obesity (90,98). Thus, we also wanted to follow research regarding the potential involvement of miRNAs in cardiovascular diseases. Indeed, also lncRNA and miRNA appear to play crucial roles in cardiovascular diseases (99,100). In particular, it has been shown that the cardiac apoptosis related lncRNA (CARL) interaction with miR-539 prevents myocyte fission and apoptosis, thereby obstructing cardiac remodeling otherwise typically following myocardial infarction (101). Studies have also reported the significance of MALAT 1 expression in endothelial cells for their proliferation during a state of hypoxia (102).
Conclusions
Various studies are suggesting that TSPO as well as ncRNAs play roles in homeostatic functions. As studies have also shown that TSPO and its ligands can regulate expression of ncRNAs, it is tempting to suggest that interactions between TSPO and ncRNAs may contribute to homeostasis of the organism. This implies that such interactions may be targeted to treat particular disorders, ranging from cancer, cardiovascular disorders, inflammatory disorders, the chronic condition of brain injury, and neurodegenerative disease.
Acknowledgments
Funding: The following funding agencies are gratefully acknowledged for their support for the research of MG, LV, and NY: KAMEA, ISF, IDF, KAMIN, EPTRI, and MAGNETON.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.12.02). JDS serves as an unpaid editorial board member of Non-coding RNA Investigation from August 2017 to July 2019. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bode J, Veenman L, Vainshtein A, et al. Modulation of Gene Expression Associated with the Cell Cycle and Tumorigenicity of Glioblastoma Cells by the 18 kDa Translocator Protein (TSPO). Austin J Pharmacol Ther 2014;2:
- Veenman L, Bode J, Gaitner M, et al. Effects of 18-kDa translocator protein knockdown on gene expression of glutamate receptors, transporters, and metabolism, and on cell viability affected by glutamate. Pharmacogenet Genomics 2012;22:606-19. [Crossref] [PubMed]
- Yasin N, Veenman L, Singh S, et al. Classical and Novel TSPO Ligands for the Mitochondrial TSPO Can Modulate Nuclear Gene Expression: Implications for Mitochondrial Retrograde Signaling. Int J Mol Sci 2017;18:786. [Crossref] [PubMed]
- Gavish M, Bachman I, Shoukrun R, et al. Enigma of the peripheral benzodiazepine receptor. Pharmacol Rev 1999;51:629-50. [PubMed]
- Gavish M, Bar-Ami S, Weizman R. The endocrine system and mitochondrial benzodiazepine receptors. Mol Cell Endocrinol 1992;88:1-13. [Crossref] [PubMed]
- Papadopoulos V, Lecanu L, Brown RC, et al. Peripheral-type benzodiazepine receptor in neurosteroid biosynthesis, neuropathology and neurological disorders. Neuroscience 2006;138:749-56. [Crossref] [PubMed]
- Veenman L, Vainshtein A, Gavish M. TSPO as a target for treatments of diseases, including neuropathological disorders. Cell Death Dis 2015;6:e1911 [Crossref] [PubMed]
- Gatliff J, East DA, Singh A, et al. A role for TSPO in mitochondrial Ca(2+) homeostasis and redox stress signaling. Cell Death Dis 2017;8:e2896 [Crossref] [PubMed]
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics 2013;12:254-78. [Crossref] [PubMed]
- Braestrup C, Squires RF. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc Natl Acad Sci U S A 1977;74:3805-9. [Crossref] [PubMed]
- Le Fur G, Vaucher N, Perrier ML, et al. Differentiation between two ligands for peripheral benzodiazepine binding sites, [3H]RO5-4864 and [3H]PK 11195, by thermodynamic studies. Life Sci 1983;33:449-57. [Crossref] [PubMed]
- Anholt RR, De Souza EB, Kuhar MJ, et al. Depletion of peripheral-type benzodiazepine receptors after hypophysectomy in rat adrenal gland and testis. Eur J Pharmacol 1985;110:41-6. [Crossref] [PubMed]
- Krueger KE, Mukhin AG, Antkiewicz-Michaluk L, et al. Purification, cloning, and expression of a peripheral-type benzodiazepine receptor. Adv Biochem Psychopharmacol 1990;46:1-13. [PubMed]
- Caballero B, Veenman L, Gavish M. Role of mitochondrial translocator protein (18 kDa) on mitochondrial- related cell death processes. Recent Pat Endocr Metab Immune Drug Discov 2013;7:86-101. [Crossref] [PubMed]
- Mukherjee S, Das SK. Translocator protein (TSPO) in breast cancer. Curr Mol Med 2012;12:443-57. [PubMed]
- Papadopoulos V, Baraldi M, Guilarte TR, et al. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci 2006;27:402-9. [Crossref] [PubMed]
- Veenman L, Gavish M, Kugler W. Apoptosis induction by erucylphosphohomocholine via the 18 kDa mitochondrial translocator protein: implications for cancer treatment. Anticancer Agents Med Chem 2014;14:559-77. [Crossref] [PubMed]
- Amhaoul H, Hamaide J, Bertoglio D, et al. Brain inflammation in a chronic epilepsy model: Evolving pattern of the translocator protein during epileptogenesis. Neurobiol Dis 2015;82:526-39. [Crossref] [PubMed]
- Cerami C, Iaccarino L, Perani D. Molecular Imaging of Neuroinflammation in Neurodegenerative Dementias: The Role of In Vivo PET Imaging. Int J Mol Sci 2017;18:E993 [Crossref] [PubMed]
- Vainshtein A, Veenman L, Shterenberg A, et al. Quinazoline-based tricyclic compounds that regulate programmed cell death, induce neuronal differentiation, and are curative in animal models for excitotoxicity and hereditary brain disease. Cell Death Discov 2015;1:15027. [Crossref] [PubMed]
- Banati RB, Middleton RJ, Chan R, et al. Positron emission tomography and functional characterization of a complete PBR/TSPO knockout. Nat Commun 2014;5:5452. [Crossref] [PubMed]
- Fan J, Campioli E, Midzak A, et al. Conditional steroidogenic cell-targeted deletion of TSPO unveils a crucial role in viability and hormone-dependent steroid formation. Proc Natl Acad Sci U S A 2015;112:7261-6. [Crossref] [PubMed]
- Li J, Papadopoulos V. Translocator protein (18 kDa) as a pharmacological target in adipocytes to regulate glucose homeostasis. Biochem Pharmacol 2015;97:99-110. [Crossref] [PubMed]
- Fan J, Lindemann P, Feuilloley MG, et al. Structural and functional evolution of the translocator protein (18 kDa). Curr Mol Med 2012;12:369-86. [PubMed]
- Veenman L, Vainshtein A, Yasin N, et al. Tetrapyrroles as Endogenous TSPO Ligands in Eukaryotes and Prokaryotes: Comparisons with Synthetic Ligands. Int J Mol Sci 2016;17:E880 [Crossref] [PubMed]
- Leneveu-Jenvrin C, Connil N, Bouffartigues E, et al. Structure-to-function relationships of bacterial translocator protein (TSPO): a focus on Pseudomonas. Front Microbiol 2014;5:631. [Crossref] [PubMed]
- Zhou ZD, Tan EK. Iron regulatory protein (IRP)-iron responsive element (IRE) signaling pathway in human neurodegenerative diseases. Mol Neurodegener 2017;12:75. [Crossref] [PubMed]
- Esmailzadeh S, Mansoori B, Mohammadi A, et al. Regulatory roles of micro-RNAs in T cell autoimmunity. Immunol Invest 2017;46:864-79. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci 2008;13:2537-47. [Crossref] [PubMed]
- Liu X, Xiao J, Zhu H, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab 2015;21:584-95. [Crossref] [PubMed]
- Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010;12:247-56. [PubMed]
- Xiao J, Bei Y, Liu J, et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J Cell Mol Med 2016;20:204-16. [Crossref] [PubMed]
- Dogini DB, Ribeiro PA, Rocha C, et al. MicroRNA expression profile in murine central nervous system development. J Mol Neurosci 2008;35:331-7. [Crossref] [PubMed]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport 2007;18:297-300. [Crossref] [PubMed]
- Lukiw WJ. Emerging amyloid beta (Ab) peptide modulators for the treatment of Alzheimer's disease (AD). Expert Opin Emerg Drugs 2008;13:255-71. [Crossref] [PubMed]
- Hébert SS, Horre K, Nicolai L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 2008;105:6415-20. [Crossref] [PubMed]
- Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci 2008;28:1213-23. [Crossref] [PubMed]
- Johnson R, Zuccato C, Belyaev ND, et al. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis 2008;29:438-45. [Crossref] [PubMed]
- Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinson’s disease. J Biotechnol 2011;152:96-101. [Crossref] [PubMed]
- Packer AN, Xing Y, Harper SQ, et al. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 2008;28:14341-6. [Crossref] [PubMed]
- Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007;317:1220-4. [Crossref] [PubMed]
- Darkazalli A, Vied C, Badger CD, et al. Human Mesenchymal Stem Cell Treatment Normalizes Cortical Gene Expression after Traumatic Brain Injury. J Neurotrauma 2017;34:204-12. [Crossref] [PubMed]
- Crick FH. The origin of the genetic code. J Mol Biol 1968;38:367-79. [Crossref] [PubMed]
- Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009;23:2639-49. [Crossref] [PubMed]
- Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014;5:171. [Crossref] [PubMed]
- Lodish H, Berk A, Zipursky SL. The Three Roles of RNA in Protein Synthesis. In: Freeman WH. editor. Molecular Cell Biology 4th edition. New York, 2000.
- Cejka D, Losert D, Wacheck V. Short interfering RNA (siRNA): tool or therapeutic? Clin Sci (Lond) 2006;110:47-58. [Crossref] [PubMed]
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494-8. [Crossref] [PubMed]
- Hammond SM, Boettcher S, Caudy AA, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001;293:1146-50. [Crossref] [PubMed]
- Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol Cell 2004;15:185-97. [Crossref] [PubMed]
- Zhou J, Shum KT, Burnett JC, et al. Nanoparticle-Based Delivery of RNAi Therapeutics: Progress and Challenges. Pharmaceuticals (Basel) 2013;6:85-107. [Crossref] [PubMed]
- Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007;129:69-82. [Crossref] [PubMed]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem 2015;84:405-33. [Crossref] [PubMed]
- Siomi MC, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011;12:246-58. [Crossref] [PubMed]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011;3:a003707 [Crossref] [PubMed]
- Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997;89:799-809. [Crossref] [PubMed]
- Yuan Y, Singh R, Reddy R. Rat nucleolar 7-2 RNA is homologous to mouse mitochondrial RNase mitochondrial RNA-processing RNA. J Biol Chem 1989;264:14835-9. [PubMed]
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci 2002;27:344-51. [Crossref] [PubMed]
- Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2010;2:a000653 [Crossref] [PubMed]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem 1995;64:897-934. [Crossref] [PubMed]
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 1990;9:2299-308. [PubMed]
- Darzacq X, Jady BE, Verheggen C, et al. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. Embo j 2002;21:2746-56. [Crossref] [PubMed]
- Deryusheva S, Gall JG. Novel small Cajal-body-specific RNAs identified in Drosophila: probing guide RNA function. RNA 2013;19:1802-14. [Crossref] [PubMed]
- Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell 2008;32:519-28. [Crossref] [PubMed]
- Kishore S, Khanna A, Zhang Z, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 2010;19:1153-64. [Crossref] [PubMed]
- Nallar SC, Kalvakolanu DV. Regulation of snoRNAs in cancer: close encounters with interferon. J Interferon Cytokine Res 2013;33:189-98. [Crossref] [PubMed]
- Cohen E, Avrahami D, Frid K, et al. Snord 3A: A Molecular Marker and Modulator of Prion Disease Progression. PLoS One 2013;8:e54433 [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci U S A 2008;105:12897-902. [Crossref] [PubMed]
- Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007;445:666-70. [Crossref] [PubMed]
- Muddashetty R, Khanam T, Kondrashov A, et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol 2002;321:433-45. [Crossref] [PubMed]
- Reeves MB, Davies AA, McSharry BP, et al. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007;316:1345-8. [Crossref] [PubMed]
- Sheik Mohamed J, Gaughwin PM, Lim B, et al. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 2010;16:324-37. [Crossref] [PubMed]
- Liu JY, Yao J, Li XM, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 2014;5:e1506 [Crossref] [PubMed]
- Puthanveetil P, Chen S, Feng B, et al. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med 2015;19:1418-25. [Crossref] [PubMed]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci 2012;13:528-41. [Crossref] [PubMed]
- Cookson W, Liang L, Abecasis G, et al. Mapping complex disease traits with global gene expression. Nat Rev Genet 2009;10:184-94. [Crossref] [PubMed]
- Mattick JS. The Genetic Signatures of Noncoding RNAs. PLOS Genetics 2009;5:e1000459 [Crossref] [PubMed]
- Stranger BE, Dermitzakis ET. From DNA to RNA to disease and back: The 'central dogma' of regulatory disease variation. Human Genomics 2006;2:383-90. [Crossref] [PubMed]
- Liu H, Wang X, Wang HD, et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat Commun 2012;3:1073. [Crossref] [PubMed]
- Funikov SY, Zatcepina OG. Mol Biol (Mosk) 2017;51:561-72. [Regulation of microRNA Activity in Stress]. [PubMed]
- O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010;33:607-19. [Crossref] [PubMed]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029-33. [Crossref] [PubMed]
- Liu GH, Zhou ZG, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 2013;34:2175-81. [Crossref] [PubMed]
- Dimitrova-Shumkovska J, Veenman L, Ristoski T, et al. Decreases in binding capacity of the mitochondrial 18 kda translocator protein accompany oxidative stress and pathological signs in rat liver after DMBA exposure. Toxicol Pathol 2010;38:957-68. [Crossref] [PubMed]
- Wang HJ, Fan J, Papadopoulos V. Translocator protein (Tspo) gene promoter-driven green fluorescent protein synthesis in transgenic mice: an in vivo model to study Tspo transcription. Cell Tissue Res 2012;350:261-75. [Crossref] [PubMed]
- Lassance L, Haghiac M, Minium J, et al. Obesity-induced down-regulation of the mitochondrial translocator protein (TSPO) impairs placental steroid production. J Clin Endocrinol Metab 2015;100:E11-8. [Crossref] [PubMed]
- Dimitrova-Shumkovska J, Veenman L, Roim I, et al. The 18 kDa Translocator Protein and Atherosclerosis in Mice Lacking Apolipoprotein E. In: Baez RV. editor. Lipoprotein metabolism: INTech, 2013:91-118.
- Thompson MM, Manning HC, Ellacott KL. Translocator protein 18 kDa (TSPO) is regulated in white and brown adipose tissue by obesity. PLoS One 2013;8:e79980 [Crossref] [PubMed]
- Li P, Ruan X, Yang L, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 2015;21:455-67. [Crossref] [PubMed]
- Sallam T, Jones MC, Gilliland T, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016;534:124-8. [Crossref] [PubMed]
- Ortega FJ, Moreno M, Mercader JM, et al. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin Epigenetics 2015;7:49. [Crossref] [PubMed]
- Karkeni E, Astier J, Tourniaire F, et al. Obesity-associated Inflammation Induces microRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function. J Clin Endocrinol Metab 2016;101:1615-26. [Crossref] [PubMed]
- Jiang X, Xue M, Fu Z, et al. Insight into the effects of adipose tissue inflammation factors on miR-378 expression and the underlying mechanism. Cell Physiol Biochem 2014;33:1778-88. [Crossref] [PubMed]
- Qi X, Xu J, Wang F, et al. Translocator protein (18 kDa): a promising therapeutic target and diagnostic tool for cardiovascular diseases. Oxid Med Cell Longev 2012;2012:162934.
- Haemmig S, Simion V, Yang D, et al. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol 2017;32:776-83. [Crossref] [PubMed]
- Kataoka M, Wang DZ. Non-Coding RNAs Including miRNAs and lncRNAs in Cardiovascular Biology and Disease. Cells 2014;3:883-98. [Crossref] [PubMed]
- Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 2014;5:3596. [PubMed]
- Lelli A, Nolan KA, Santambrogio S, et al. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia 2015;3:45-52. [PubMed]
Cite this article as: Yasin N, Veenman L, Dimitrova-Shumkovska J, Gavish M. The 18 kDa translocator protein, non-coding RNA, and homeostasis. Non-coding RNA Investig 2017;1:25.