Argonaute-crosslinking and immunoprecipitation deciphers the liver miR-122 targetome
The profiling of microRNA (miRNA) expression in the liver by different groups has revealed that miR-122 is one of the most abundant miRNAs in this organ, accounting for 70% of miRNA expression in the liver (1-4). A systematic study of the miRNA patterns of different tissues in mice also demonstrated that miR-122 was a tissue-specific miRNA, highly enriched in liver tissue but absent from other tissues (5). The miR-122 sequence is completely conserved in vertebrates and no orthologues have been detected either in D. melanogaster or in C. elegans, indicating that miR-122 is probably linked to the appearance of the liver in vertebrates (6). In this line, the described functions of miR-122 are closely related to the regulation of specific hepatic processes, including cholesterol and lipid metabolism (7,8). Interestingly, miR-122 has been demonstrated to play a role in the regulation of hepatic polyploidy (9), a defining feature of the liver, and is crucial for hepatitis C virus replication in the liver (10). During carcinogenesis, miR-122 becomes downregulated and its expression levels are indicative of patient outcome in hepatocellular carcinoma (HCC) (11). Moreover, two miR-122 Knockout (KO) mice models, germline KO and liver KO, showed that inhibition of miR-122 activated an inflammatory process in the liver that led to hepatitis in the short term and to HCC in the long term (12). In contrast, the delivery of miR-122 to a MYC-driven mouse model of HCC strongly inhibited tumorigenesis, reinforcing the role of miR-122 as a tumor suppressor gene (12). Although several targets associated with previously described functions of miR-122 have been identified, the miR-122 targetome in normal liver and in HCC remained incomplete until now.
The most frequent strategy for miRNA target identification is usually based on bioinformatics through different computational algorithms followed by an experimental validation through luciferase reporter assays and/or western blot analysis after intervention of miRNA levels (13). Most of the computational algorithms, including the three most commonly used, TargetScan, miRanda and PicTar, are based on the identification of sequence complementarities between miRNAs and the 3'UTR regions of putative target genes according to different rules. Usually these rules include the use of the 2 to 8 NT from the 5'UTR region of the miRNA, known as seed sequence. The seed sequence is known to be highly conserved and in most cases the matching of the seed sequence with the 3'UTR region is sufficient to confer mRNA recognition (14). A few prediction tools, such as miRcode, also take into account coding regions (CDS) or 5'UTR regions, but this is less common. Moreover, prediction tools usually ignore the transcriptome information of cell types under study and the non-canonical binding sites. Due to these factors, many predicted targets are finally not experimentally validated and many real targets can be missed since the false positive rate for these prediction tools ranges from 24% to 70% (15,16).
In contrast, new methods have emerged in recent years that allow large-scale miRNA target identification to identify the complex networks regulated by one miRNA (17). Most of these methods are based on the overexpression or inhibition of the miRNA of interest by different methods followed by a downstream gene-expression or proteomic analysis. Among these methods, argonaute-crosslinking and immunoprecipitation (AGO-CLIP, also known as high-throughput sequencing of RNA isolated by CLIP, HITS-CLIP, or CLIP-seq) followed by deep sequencing allows the mapping of miRNA binding to AGO sites and can provide a single base-pair resolution of miRNA and AGO binding sites (18,19) (Figure 1). However, this technique has two limitations: the low efficiency of UV254 nm RNA-protein crosslinking (19) and the fact that it can only identify about 100 NT around the specific target site (18). Therefore, the sites identified by AGO-CLIP will need a validation by a different technique to verify the identified target sites.
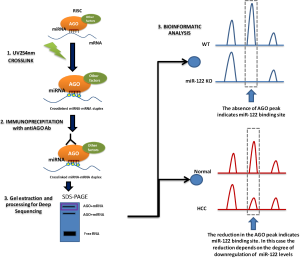
Recently, Luna et al. (20), using the AGO-Clip technique, defined the miR-122 targetome in mouse and human liver with the identification of more than 11,000 genomic loci displaying clear miR-122 binding dependence. The significance of the first part of the study lies in the use of the miR-122 KO mice previously generated by the group (12). Performing AGO-CLIP in miR-122 KO and in miR-122 floxed [wild type (WT)] mice is an ideal platform for clarifying the liver miR-122 targetome in vivo. In this model, the authors evaluated the absence of Ago binding peaks in the KO samples (Figure 1), which provided evidence that miR-122 engages a larger fraction of the transcriptome than previously assumed. The study confirmed previous reports that miR-122 was one of the most frequent miRNAs in liver and—together with let-7, miR-15/16, miR-17, miR-21a, miR-26, miR-27, miR-30, miR-192, and miR-194—conformed the top ten miRNA families in mouse liver. Remarkably, miR-122 represented 18.7% of binding events of the top ten miRNAs (44% of all binding events). The AGO-CLIP analysis identified 9,280 miR-122 canonical binding sites, which were located not only in the 3'UTR region of the target genes (~30%) but also in CDS (~27%), intronic (~23%) and 5'UTR (~4%) regions. Moreover, the motif analysis also showed 1,923 non-canonical, G-bulged, binding sites, which more frequently were found associated to CDS regions.
The authors then evaluated how these results correlated with the gene expression patterns (RNAseq data) observed in the KO or the WT mice. Despite the dramatic reduction of ago binding in KO livers, only 29% of the targets with 3'UTR or CDS binding sites and 15% of targets with 5'UTR or intronic binding sites were significantly upregulated in KO livers. Since miRNAs mostly affect the translation rates of its targets and only affect mRNA levels in a small proportion of targets, the authors would probably have obtained higher percentages using proteomic analysis rather than expression analysis. Moreover, several consequences of miRNA-target interactions beyond repression of gene expression or translation have been identified that could also explain the low percentage of targets with increased expression in the KO mice (21). However, the greatest changes in gene expression were associated with downstream genes of the miR-122 targets, with 1,600 transcripts upregulated and 1,300 downregulated in KO livers. Pathway analysis showed that the most relevant signaling pathways involved included the “cell-cell junctions” and “cytoskeletal rearrangements” pathways. Both pathways have previously been demonstrated to play a significant role in the proliferation regulation of liver cells (22).
In the second part of the work, the authors completed the study of the miR-122 targetome by studying tumor and paired normal tissue from nine patients with HCC. The AGO-CLIP data confirmed that miR-122 was significantly downregulated in tumor tissue and that both canonical and G-bulged miR-122 3'UTR sites were significantly less bound in tumor samples, while oncogenic miRNAs such as miR-21 were more bound. This was also observed in CDS, introns and 5'UTR. In contrast with the KO mouse model, where the presence or absence of miR-122 allowed the clear identification of directed targets, in HCC samples the AGO/CLIP can be affected by the grade of downregulation of miR-122 (Figure 1). In fact, the authors observed that the mean CLIP signal for miR-122 binding was highly correlated with tumor miR-122 levels, indicating the existence of a deregulated miR-122 targetome in human HCC.
The comparison of the list of targets obtained in the two models (KO mouse and HCC) revealed that the miR-122 targetome has an unusually low conservation. When the authors compared the 3'UTR and CDS targets in the human HCC samples with those in the KO mouse model, they observed only a 20% overlap between the species. Specifically, of the 4,771 identified targets in the two models, only 965 were shared. This was in contrast with the higher conservation of the targetome of the other miRNAs included in the top ten and also with the general thinking that links miRNA conservation and target site conservation with an effective target repression (14). The authors suggest that this may be due to the fact that miR-122 is not crucial for liver development and thus the gain or loss of miR-122 binding sites is relatively neutral. These results highlight the fact that some bioinformatics tools for target prediction emphasizing conservation can fail with some miRNAs, such as miR-122, where 76% of mouse and 45% of human targets were species-specific. Curiously, several of the shared targets have functions in maintaining normal liver homeostasis and the authors selected these targets (GNPDA2, ZBTB4, CDA, GPR107, ARHGAP1, ZMIZ1, SNAI2, Cept1 and Sp2) for further validation by luciferase assay and quantitative polymerase chain reaction (qPCR) and verified that they had functional binding sites. Additionally, the shared targets also belong to cell cycle and tight junction pathways, as well to cancer pathways (e.g., AMPK, PI3K/AKT, and WNT/β-catenin).
Further analysis of the 965 shared targets showed that they could have clinical implications in HCC, since some correlated with patient survival according to data from the Cancer Genome Atlas (TCGA Research Network), which included 373 usable cases of HCC. The authors hypothesized that the list of 965 shared targets of miR-122 would be upregulated in tumor tissue, where miR-122 is downregulated, compared to normal tissue, where miR-122 is upregulated. However, although they observed a general upregulation in the LIHC cohort of the majority of these targets, their heatmap shows different clusters of patients, including a small one with general downregulation of the targetome. The authors did not perform additional clinical correlations that could explain these inter-patient variabilities. However, the authors identified a group of three upregulated targets, BCL2, SLC52A2, and STX6, whose expression levels correlated with survival in the LIHC cohort. To verify that the upregulation of these three genes was not limited to the TCGA data, the authors confirmed their upregulation in two additional RNAseq datasets from GEO: GSE77509 (n=20) and GSE77314 (n=50). The expression of these upregulated genes negatively correlated with miR-122 expression and their high levels were significantly associated with poor survival, both alone and in combination. Only BCL9 had previously been associated with survival in HCC; a Korean study of BCL9 protein expression, evaluated by immunohistochemistry in 288 HCC patients, found that the 74 patients with nuclear BCL9 protein expression had shorter disease-free survival (23).
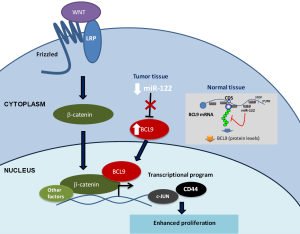
After validation by luciferase and qPCR of the miR-122 regulation of these three targets, the authors focused on the functional implications of miR-122 regulation of BCL9, a WNT pathway member that coactivates β-catenin (24). The authors chose to focus on BCL9 because the WNT pathway is critical to HCC and because BCL9 harbors several binding sites in its CDS region—six in humans and seven in mice—indicating that it is highly regulated by miR-122. To verify the relevance of these binding sites, the authors performed luciferase reporter assays that showed that 5/7 mouse and 3/6 human binding sites mediated target repression. Moreover, using an FLAG-tagged human BCL9 and miR-122 mimic in H293T cells, the authors showed that protein and mRNA levels of BCL9 were suppressed by miR-122 but not when the binding sites were mutated. The study of the functional repercussions of the miR-122-BCL9 regulation showed that miR-122 regulated β-catenin activation and impacted the β-catenin transcriptional program by inhibiting BCL9 (Figure 2). Cells overexpressing BCL9-WT or BCL9 with mutated miR-122 binding sites (BCL9-MUT) increased β-catenin activity. However, only BCL9-WT cells treated with miR-122 mimic suppressed β-catenin activity. The BCL9 knockdown in MHCC-LM3 cells showed that β-catenin response elements CD44 and c-JUN saw reduced their expression levels. Moreover, BCL9 overexpression in two cell lines (MHCC-LM3 and Huh7.5) was associated with increased proliferation. BCL9 is a conserved miR-122 CDS target that may impact WNT-mediated progression of HCC specifically through proliferation regulation (Figure 2).
In summary, Luna et al. have identified greater than 104 distinct target loci across genic and non-genic regions, including canonical and non-canonical bindings for miR-122. Interestingly, they observed that the changes in gene expression were linked mostly to 3'UTR and CDS targeting and that most of the miR-122 targets were species-specific. Furthermore, among the 965 targets shared between humans and mice, which were mostly upregulated in HCC tumor samples, BCL9, SLC52A2, and STX6 correlated with patient survival. Finally, miR-122 was able to regulate the crucial WNT/β-catenin pathway in HCC through BCL9.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.11.08). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gamazon ER, Innocenti F, Wei R, et al. A genome-wide integrative study of microRNAs in human liver. BMC genomics 2013;14:395. [Crossref] [PubMed]
- Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer cell 2011;19:232-43. [Crossref] [PubMed]
- Law PT, Qin H, Ching AK, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol 2013;58:1165-73. [Crossref] [PubMed]
- Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci 2016;17:280. [Crossref] [PubMed]
- Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12:735-9. [Crossref] [PubMed]
- Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol 2012;9:137-42. [Crossref] [PubMed]
- Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87-98. [Crossref] [PubMed]
- Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 2005;438:685-9. [Crossref] [PubMed]
- Hsu SH, Delgado ER, Otero PA, et al. MicroRNA‐122 regulates polyploidization in the murine liver. Hepatology 2016;64:599-615. [Crossref] [PubMed]
- Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005;309:1577-81. [Crossref] [PubMed]
- Coulouarn C, Factor VM, Andersen JB, et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009;28:3526-36. [Crossref] [PubMed]
- Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 2012;122:2871-83. [Crossref] [PubMed]
- Kuhn DE, Martin MM, Feldman DS, et al. Experimental validation of miRNA targets. Methods 2008;44:47-54. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett 2005;579:5904-10. [Crossref] [PubMed]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods 2006;3:881-6. [Crossref] [PubMed]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res 2011;39:6845-53. [Crossref] [PubMed]
- Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479-86. [PubMed]
- Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129-41. [Crossref] [PubMed]
- Luna JM, Barajas JM, Teng KY, et al. Argonaute CLIP Defines a Deregulated miR-122-Bound Transcriptome that Correlates with Patient Survival in Human Liver Cancer. Mol Cell 2017;67:400-10.e7. [Crossref] [PubMed]
- Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions [mdash] beyond repression of gene expression. Nat Rev Genet 2014;15:599-612. [Crossref] [PubMed]
- Tsukita S, Yamazaki Y, Katsuno T, et al. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 2008;27:6930-8. [Crossref] [PubMed]
- Hyeon J, Ahn S, Lee JJ, et al. Prognostic Significance of BCL9 Expression in Hepatocellular Carcinoma. Korean J Pathol 2013;47:130-6. [Crossref] [PubMed]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17:9-26. [Crossref] [PubMed]
Cite this article as: Navarro A. Argonaute-crosslinking and immunoprecipitation deciphers the liver miR-122 targetome. Non-coding RNA Investig 2017;1:23.