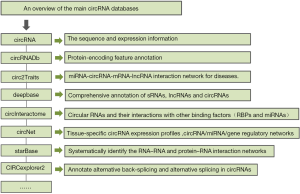
An overview of the main circRNA databases
Introduction
Unlike linear RNA, circular RNA (circRNA) is a group of non-coding RNA which forms a covalently closed continuous loop from exon circularization. The first circRNA was identified in 1979, when the researcher revealed the fact that RNAs could exist in circular form in the cytoplasm of eukaryotic cells (1). Ten years later, very low levels of transcripts of the deleted colorectal carcinoma (DCC) gene with scrambled exons in human cytoplasmic RNA were reported (2). Because of the low expression level and special structure, only a few genes were identified to express circRNAs, such as DCC and sex-determining region Y (SRY). In recent years, high throughput sequencing technology has dramatically expanded the scope of transcriptomics research (3) and a large number of circRNAs have been discovered across species (4-7). Recently, thousands of circRNAs expressed in animal cells (8-12) have been reported, and the numbers are likely to grow.
These circRNAs are unusually stable RNA molecules, presumably because their lack of ends prevents them from regulation by conventional RNA degradation pathways. Moreover, they are found to be evolutionary conservative, and specifically expressed across tissues or developmental stages, and play important roles in gene regulation (11).
The functional roles of circRNAs are not as well unknown as those of other noncoding RNAs such as microRNAs (miRNAs). However, one popular mechanism whereby circRNAs are believed to function is by sponging miRNAs, sequestering them away from protein-coding mRNAs (12). Owing to their ability for sequestration of miRNAs, circRNAs play an important role in fine tuning of post-transcriptional gene expression. Their interaction with disease associated miRNAs indicates that circRNAs dysregulation is responsible for the genesis of many diseases. In addition, circRNAs can also be interesting and novel class of biomarkers.
Therefore, circRNA has become the hotspots in the current transcriptomics research field.
To enable the study of circRNAs, several databases have been provided such as circBase, circRNADb, circ2traits, deepBase, circInteractome and circNet (13-19). Here we have described these databases and their features respectively, which are summarized in Figure 1.
The database circBase (13) contains data from all studies of large-scale circRNA identification published up to 2013 and is regularly updated with newly published data (20,21). It is developed by Glažar P et al. and can be freely accessible through the website at http://www.circbase.org/.
As a database merged and unified data sets of circRNAs from public references, the database currently hosts data from various Homo sapiens, Mus musculus, Caenorhabditis elegans, and Latimeria samples. However, circBase does not cover viroids, which are already collected in other resources. The circRNA sequence, as well as the evidence supporting their expression can be accessed, downloaded, and browsed within the genomic context. Besides, circBase also provides scripts to identify known and novel circRNAs in sequencing data.
There are three main ways to query circBase: (I) simple search, available from the server homepage, is intended for simple queries by identifiers, genomic location, sequence, gene description, or Gene Ontology term identifiers; (II) list search, allows the intersection of a large number of search terms with database contents. Upon selecting the organism and assembly, the user can paste or upload a list of any identifier type supported by circBase. It is also possible to submit a list of genomic regions (for example in BED format) and retrieve all overlapping circRNAs in the database; (III) table browser can be used for conditional data retrieval. After selecting the organism and experiment of interest, the user can further refine the selection by a number of options, such as presence in a particular sample, range of genomic or spliced sequence lengths, number of reads supporting the head-to-tail splice junction, and many others.
They intend to regularly update the database with newly published data. Direct data submission by the users is currently not supported, but the users are encouraged to contact them with requests for adding their data to circBase.
Circ2Traits (14), a comprehensive database for circRNAs potentially associated with diseases in human, is compiled by Suman Ghosal et al. and accessible through the website at http://gyanxet- beta.com/circdb/.
The present version of circ2Traits has categorized 1951 human circRNAs potentially associated with 105 different diseases. Furthermore, circ2Traits stores the complete putative miRNA-circRNA -mRNA-lncRNA interaction network for each of these diseases. CircRNAs and their information stored in Circ2Traits are categorized according to their potential association with diseases, as observed from the GWAS associated SNPs and potential interaction with disease associated miRNAs.
There are several search options in this database. To begin with, the user can choose from a directory of the 105 diseases, to view a list of circRNAs most likely to be associated with the disease and also visualize the interaction network and see the interaction table for each disease. There are other search options like keywords based search for miRNAs, circRNAs, protein coding genes (symbols or mRNA accession), and lncRNAs. Search options for GWAS traits associated circRNAs are also available. For each circRNA, information like traits associated and other SNPs, Ago interaction sites are stored besides general information about the circRNA (name, locus, and interacting miRNAs). The information on individual circRNA can be viewed upon selecting a circRNA from the interaction table.
In the future versions of the database, more elaborate study of the binding sites for the different RNA-binding proteins in the circRNA loci will be incorporated. As the field matures, and the expression levels of circRNAs become more precisely known, the developers hope to include the tissue specific expression patterns of the circRNAs to make it a more comprehensive and useful tool.
CircNet (15), a database constructed by transcriptome sequencing datasets, is developed by Yu Chen Liu et al. The web tool is freely accessible at http://circnet.mbc.nctu.edu.tw/.
In circNet, previously reported and newly identified human circRNAs are cataloged, with circRNA expression metadata in the form of a heatmap illustrating circRNA expression profiles across 464 human transcriptome samples additionally provided. It provides tissue-specific circRNA expression profiles and circRNA-miRNA-gene regulatory networks. It not only extends the most up to date catalog of circRNAs but also provides a thorough expression analysis of both previously reported and novel circRNAs. Furthermore, it generates an integrated regulatory network that illustrates the regulation between circRNAs, miRNAs and genes.
Overall, circNet provides interactive tools for users to easily access comprehensive information regarding expression profiles across many conditions, genome loci, close repeat sequences, post-transcription regulation networks, and references to previous studies for circRNAs. In circNet, users can choose an interested gene or miRNA as the keyword, then, available information including the integrated mRNA-miRNA-circRNA regulatory network, expression profile, and genome position were collected.
CircNet would be a useful tool to study circRNA tissue specific function as well as correlation to disease. Further inquiries using the regulatory networks identified using CircNet may discover additional novel feedback loops with applicability to human disease.
CircInteractome (16) is developed by Dawood B. Dudekulay et al. The web tool is freely accessible at http://circinteractome.nia.nih.gov, facilitating the analysis of circRNAs and their interactions with other binding factors, mainly RBPs and miRNAs.
CircInteractome provides researchers with valuable details about circRNAs and their possible role in sequestering RBPs and/or miRNAs and thereby reduces their availability for mRNAs. CircInteractome also facilitates the design of primers for studying circRNA by RT-Qpcr analysis. CircInteractome can be used to predict RBP binding to upstream and downstream sequences of the pre-spliced transcript, thus potentially shedding light into the biogenesis of circRNAs. The interaction of translation regulators (miRNAs, RBPs, ITAFs) and the presence of internal ribosome entry sites (IRESs) in circRNAs can help to decipher whether circRNAs have protein/peptide-coding capabilities.
CircInteractome incorporates several features from other freely available web resources, such as circBase, StareBase 2.0, TargetScan 7.0, and Primer3. By integrating these resources, CircInteractome enables the user to find out the genomic and mature circRNA sequences, circRNA-binding partners (RBPs, miRNAs), siRNAs, and primers to study circRNA levels, localization, and function.
Although CircInteractome has many user-friendly features for circRNA researchers, it is limited in its ability to predict RBP and miRNA interactions when circRNAs form secondary or tertiary structures. Given that all of the data provided here are predicted based on sequence matches and the presence of secondary or tertiary structures in circRNA cannot be considered systematically, experimental validation is essential to verify RBP and miRNA functional sites.
The explorers plan to maintain, update and curate CircInteractome in the foreseeable future, and will include additional RBPs, miRNAs, and circRNAs as they become available. Secondary structure, experimental validation of circRNAs interacting with RBPs and miRNAs will also be included in CircInteractome. They will also integrate circRNAs from other species such as mouse and monkey, and will include predictions of RNA hybrids including circRNA:mRNA and circRNA:lncRNA.
CircRNA (17) database, also known as circRNADb, contains 32,914 human exonic circRNAs which were carefully selected from diversified sources by Xiaoping Chen et al. The database is freely accessible at http://reprod.njmu.edu.cn/circrnadb.
CircRNADb provides the detailed information of the circRNA, including genomic information, exon splicing, genome sequence, IRES, open reading frame (ORF) and references. As a comprehensive human exonic circRNA database with protein-encoding feature annotation, circRNADb is designed to provide a rich data resource for circRNAs research. circRNADb has collected circRNA dataset from relevant literatures and the brain RNA-seq dataset from the authors’ work. In total, 32,914 non-redundant human exonic circRNAs are obtained. CircRNADb may facilitate circRNA studies by (I) providing users with detailed genomic information of each circRNA; (II) annotating protein-coding potential of each circRNA; (III) including protein expression evidences of circRNA by mass spectrometry; (IV) providing convenient interfaces to retrieve the data.
Users can enter the search terms in the search text box, such as chromosome name, gene symbol, transcript, and other keywords to query the circRNA, then the results that matched the query keywords will be listed in the result page. The dataset in circRNADb can be browsed in three options: gene symbol, PubMed ID, and cell type. The detailed information section provides the best matched transcript of the circRNA, its exons information and spliced sequences. According to gene annotation GTF file, all possible circular isoforms and related information are also displayed. If the circRNA has the potential to code a protein, protein features including domains, post-translational modification sites and half-life prediction are provided. In addition, we annotated circRNA parental gene associated with human disease (OMIM).
Collectively, this database provides the function of data search, browse, download, submit and feedback for the user to study particular circRNA of interest and update the database continually. CircRNADb will be built to be a biological information platform for circRNA molecules and related biological functions in the future.
The deepBase v2.0 (18) facilitates the integrative, interactive and versatile display of, as well as the comprehensive annotation and discovery of sRNAs, lncRNAs and circRNAs, which is freely available at http://deepbase.sysu.edu.cn/ or http://biocenter.sysu.edu.cn/deepBase/.
The deepBase v2.0 was described as an updated platform, to decode evolution, expression patterns and functions of diverse ncRNAs across 19 species. For circRNAs they annotated 14,867 human circRNAs, and 1,260 of which are orthologous to mouse circRNAs.
The distinctive features of deepBase v2.0 include the following: (I) providing the comprehensive expression analysis of sRNAs and lncRNAs from 1,036 RNA-Seq datasets from 19 species. The constructed gene expression profiles of both ncRNAs and protein-coding genes are valuable for understanding the similarities and differences of transcriptional regulation of protein-coding genes and ncRNAs across different tissue/cell-line types; (II) constructing evolutional patterns of lncRNAs and circRNAs across several evolutional clades. Conservation patterns may help biologists to select important ncRNAs for further functional validation; (III) two web-based tools, lncSeeker and lncFunction, can be used to identify high-confidence lncRNAs, and to predict lncRNA functions.
StarBase v2.0 (19) (http://starbase.sysu.edu.cn/) is developed by Jun Hao Li et al. with the ability to systematically identify the RNA–RNA and protein-RNA interaction networks from 108 CLIP-Seq (PAR-CLIP, HITS-CLIP, iCLIP, CLASH) data sets generated by 37 independent studies.
By analyzing millions of RNA-binding protein binding sites, 9,000 miRNA-circRNA, 16,000 miRNA pseudo gene and 285,000 protein-RNA regulatory relationships were identified. Moreover, starBase v2.0 has been updated to provide the most comprehensive CLIP-Seq experimentally supported miRNA-mRNA and miRNA-lncRNA interaction networks to date. Extensive and complex RNA-RNA and protein-RNA interaction networks have been shown by analyzing a large set of Ago and RBP binding sites derived from all available CLIP-Seq experimental techniques.
Compared with other databases, the distinctive features of starBase v2.0 include the following: (I) providing the miRNA-pseudogene interaction networks; (II) drafting the interaction maps between miRNAs and circRNAs; (III) providing an enhanced resolution to determine ceRNA functional networks based on miRNA-target interactions overlapping with high-throughput CLIP-Seq data; (IV) providing comprehensive miRNA-lncRNA interactions; and (V) providing a variety of interfaces and graphic visualizations to facilitate analysis of the massive and heterogeneous CLIP-Seq, RBP binding sites, miRNA targets and ceRNA regulatory networks in normal tissues and cancer cells.
CIRCexplorer2 is developed by Chen LL, Yang L et al., which is a comprehensive and integrative database, aiming to annotating alternative back-splicing and alternative splicing in circRNAs across different cell lines. It is freely available at http://circexplorer2.readthedocs.io/en/latest/.
It is the successor of CIRCexplorer with plenty of new features to facilitate circRNA identification and characterization. The distinctive features of CIRCexplorer2 include the following: (I) support multiple circRNA aligners; (II) de novo assemble novel circRNA transcripts; (III) characterize various alternative (back-) splicing events of circRNAs; (IV) fast identify circRNAs with STAR or BWA; (V) support both single-read and paired-end sequencing.
Conclusions
In recent years, with the development of high throughput sequencing technology, a large number of circRNAs have been discovered across species, and the numbers are likely to grow. Next generation sequencing technologies also play vital roles in improving our understanding of functional genomics. So far, several databases of the circRNA have been published with different features. We believed that in the future versions of the existed database and newly merged databases, more specific and comprehensive information will be contained to help revealed the molecular mechanism and biological functions.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.11.05). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. [Crossref] [PubMed]
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57-63. [Crossref] [PubMed]
- Danan M, Schwartz S, Edelheit S, et al. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 2012;40:3131-42. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666-70. [Crossref] [PubMed]
- Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013;4:283. [Crossref] [PubMed]
- Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016;44:D209-15. [Crossref] [PubMed]
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016;13:34-42. [Crossref] [PubMed]
- Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 2016;6:34985. [Crossref] [PubMed]
- Zheng LL, Li JH, Wu J, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 2016;44:D196-202. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
Cite this article as: Xu Y. An overview of the main circRNA databases. Non-coding RNA Investig 2017;1:22.