
The multifaceted regulatory potential of tRNA-derived fragments
tRNAs have been discovered in the late 1950-ies of the last century and have subsequently been characterized biochemically, genetically and structurally as amino-acid delivering molecules for protein biosynthesis. First reports of stable tRNA-derived cleavage products have been reported in the late 1970-ies (1), but at that time they were mostly regarded as functionless degradation products. The advent of high-throughput sequencing techniques in the last years allowed the discovery of an unforeseen variety of small non-protein-coding RNAs (ncRNAs), including tRNA-derived RNA fragments (typically referred to as tdRs or tRFs), in all domains of life. Post-transcriptional RNA cleavage encompasses all sorts of transcripts including mRNAs as well as ncRNAs thus amplifying the biological repertoire of cellular RNomes significantly. Most of these small ncRNAs are involved in different steps of gene regulation. The most studied and well understood small ncRNA regulators are the ~22 nt long miRNA and siRNA involved in postranscriptional gene silencing, a process termed RNA interference (RNAi). Mi/siRNAs are present in almost all eukaryal cells and they regulate mRNA stability and/or translatability by utilizing sequence complementarity to their mRNA targets. tRNA-derived RNAs (tdRs) have meanwhile also made it on the ever growing list of ribo-regulators. tdRs, which include tRNA halves (size 30–35 nt) as well as shorter fragments (~14–26 nt), were first discovered in cells exposed to different stress conditions [in E. coli infected with T4 virus (2); in Tetrahymena under amino acid starvation (3), in human cells (4)], but further studies have shown that they are also constitutively expressed in several cells and tissues.
The biological function(s) of tdRs remained largely elusive until 2009, when it was shown that tdRs originated by cleavage of tRNAs in the anticodon loop were able to repress translation during stress (5). In vast contrast to mi/siRNAs, that more or less all follow the same biogenesis in all species and, more importantly, execute their biological function in a highly homologous manner, tdRs are functionally extremely heterogeneous. First, they are not restricted to one domain of life since they have been found in archaea, bacteria as well as in uni- and multicellular eukarya. Second, the biological roles of tdRs embrace regulation of transcription (6), translation (5,7-9), stress granule formation (10), apoptosis (11), cell proliferation (12,13), RNAi (14), vesicle-mediated intercellular communication (6), and intergenerational inheritance (15) (Figure 1). Therefore, the mere detection of a tRNA fragment in a deep-sequencing screen does not allow reasonable deductions to be made about its biogenesis or function. This on the other hand means that basically each and every tRNA-derived fragment needs to be individually investigated by dedicated mechanistic studies.
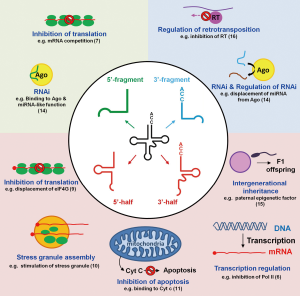
The paper published recently in Cell by Schorn et al. is a good example of such a focused tdR study (16). The authors open yet another chapter about the regulatory repertoire of tdRs. In this publication, they describe a so far unprecedented function of tdRs, namely as safeguards of genome integrity in mammalian cells. Retroviruses and retrotransposons are mobile elements present in many genomes that depend on unique enzymes, namely reverse transcriptase and transposase for their propagation. Due to their ability to “jump” to different locations they pose a threat for potential germline mutations and therefore need to be tightly regulated. To keep them under control cells have developed a variety of strategies. Typically transcription of transposable elements is regulated epigenetically by DNA methylation (performed by DNMT1) and histone H3K9 trimethylation by SETDB-1. Under certain circumstances though, cells need to achieve pluripotency and therefore undergo epigenetic reprogramming. When this happens the epigenetic markers that control transposable element mobility are also removed. To nevertheless prevent genome damage transposable elements are kept under control by means of other pathways that involve two families of small ncRNAs: piwi-interacting RNAs (piRNAs) and endogenous small interfering RNAs (endo-siRNAs). piRNAs act mainly in the germline while endo-siRNAs inactivate transposons immediately after fertilization (17). Until now there was no information about the regulatory mechanisms that prevent transposable element mobility during early embryo development, before the epigenetic marks are re-established. Schorn et al. discovered a novel mechanism how tdRs are able to repress the mobility of long terminal repeat (LTR)-retrotransposons in preimplantation stem cells in mouse (16). Screening for small RNAs putatively involved in the control of transposable elements they could identify 3'-tdRs of 18- and 22-nt length, all containing the universally conserved 3'-CCA end of mature tRNAs. These tdRs show clear complementarity to specific regions of retrotransposons. The 18 nt tdRs identified match with almost perfect complementarity to the 5' UTR of the LTR-retrotransposon’s genomic sequence exactly at the highly conserved primer binding site (PBS). The PBS is an essential region of the primary retrotransposon transcript required for annealing of cellular full-length tRNAs to prime reverse transcription for cDNA synthesis. The authors could convincingly demonstrate that the 18 nt long tdR inhibited retrotransposition of the two most active mouse transposons using a plasmid-based retrotransposition assay in cell culture. This inhibition of transposition was not due to changes in the transcription levels as there were no apparent changes in the levels of histone methylation at relevant loci. The authors could also discard the possibility of an RNAi-like function of the 18-mer tdR, since no effects on the RNA stability and on the translational efficiency of retrotransposon mRNAs were observed. On the other hand, deep-sequencing of RNaseH cleavage intermediates, canonical processing products of the retroviral transcript that depends on the successful production of tRNA-primed cDNA in the regular life cycle, showed an inhibitory effect of tdRs on reverse transcriptase activity. In their mechanistic model, the authors suggest that the 18-mer tdR interferes with reverse transcription by hybridizing to the PBS and directly competing with the regular full-length tRNA usually utilized as primer for reverse transcription (Figure 1). The paper by Schorn et al. is not the first report of tdRs being possibly involved in regulating the retroviral life cycle. As early as in 2009 Yeung et al. identified a hybrid formed by an 18-mer tdR and the PBS of HIV-1 (18). In 2014 Ruggero and colleagues discovered a 3'-tdR originating from tRNAPro of again 18-nt length fully complementary to the PBS of human T-cell leukemia virus HTLV-1 (19). In striking contrast to the study of Schorn et al., this 18-mer tdR was proposed to stimulate priming of revere transcription in an in vitro reverse transcription assay. This example most impressively highlights the functional versatility of tdR biology: in both cases an 18-mer tdR shows perfect complementarity to the PBS of a retroviral transcript, yet one inhibits (16) and the other stimulates (19) cDNA production. In the study published in Cell by Schorn et al., the second most abundant tdRs were 22-nt long (16). This class of tdR differs from the above described 18-mer fragment only by four additional nucleotides at the 5' end. The 22-mer tdR, in contrast, seem to behave miRNA-like and has been shown to participate in regulating transposable elements at the translational level in retrotransposons that still carry the PBS (16).
Several important questions still await experimental confirmation in the proposed mechanism. One of the most urging ones is how are the two different species of 3'-tdRs, the 18-mer and the 22-mer, produced? Is it the same enzyme or are different biogenesis pathways involved? Similar to the plethora of tdRs functional diversity, also the biogenesis of the various tdRs observed throughout different model system seem to be extremely heterogeneous. While it is well established that angiogenin, a small ribonuclease of the RNAse A family, is responsible for the production of stress-induced tRNA halves in mammalian cells (5,9), other eukarya that lack angiogenin orthologs are still capable of producing tRNA halves upon adverse environmental conditions (e.g., in Trypanosoma: (20) and our unpublished data). Homologues of RNase T2 (e.g., Rny1) are responsible for the production of tRNA halves in yeast and possibly also in plants. Other enzymes that have been implicated in 5'- or 3'-tdR processing in pro- and eukaryal species involve dicer, and colicin D & E5, to name a few (21,22). Another pending question to be addressed concerns the role of modified nucleosides in tRNA molecules for tdR biogenesis. tRNAs are known to carry numerous post-transcriptionally added nucleobase and ribose modifications that are predominately located in and around the tRNA anticodon and the D- and the T-loops. It has been shown in some cases that indeed the presence of a particular tRNA modification affects the production of tdRs (23). The 22-mer tdR identified in Schorn et al. could potentially contain two modified nucleosides at the 5'-end remaining from the corresponding T-loop modifications of the mature tRNALys, which was however not experimentally demonstrated in this publication (16). The 18-mer tdR that differs from the former by lacking the four utmost 5' nucleosides are expected to be completely unmodified. It is tempting to speculate that these differences in the RNA modification pattern are involved in establishing the markedly different regulatory functions of these tdRs: the 22-mer tdR resembles miRNAs and inhibits translation while the 18-mer tdR interferes with reverse transcription in an RNAi-independent manner (16). Comprehensive experimental focus on the role of modified nucleosides for tdR function as well as for tdR biogenesis is currently largely lacking in the field. It is conceivable that the still poorly understood and largely enigmatic biological roles of tRNA modification need to be re-evaluated in the light of tdR biology.
The pivotal role modifications play in tdR biology can be better exemplified by studies on human diseases. It has been shown that lack of tRNA methylation positively affects the production of tdRs, and this in turn correlates with the severity of certain neurological disorders (23,24). These data highlight a possible causal connection between the tRNA-modification pattern, tdR biogenesis as well as abundance, and disease progression. The connection between tdRs and human disease is not limited to neurodevelopmental disorders. Many studies have identified tdRs as major players in different types of cancer, affecting cancer malignancy and progression (for a recent review see ref. 25).
It is surprising that the “mother molecule” of tdRs, genuine tRNA that is, has basically one major cellular role in targeting and delivering amino acids to translating ribosomes, while processing products thereof are functionally so multifaceted. Introduction of a single cleavage at a particular phosphodiester bond of a tRNA molecule thus can give birth to smaller RNA entities with completely different and unpredictable biological roles. Reports on this growing family of tRNA-derived small non-coding RNAs are accumulating in recent years and according to the tRFdb (http://genome.bioch.virginia.edu/trfdb/) it already comprises more than 2,500 members.
In summary, tdR research has undergone dramatic changes over the past 40 years: originally considered meaningless degradation products of tRNA, tdRs have proven to be crucial players in orchestrating gene regulation in health and disease. As the list of newly identified tdR sequence entries expands rapidly, so will hopefully the number of dedicated functional analyses for this incredibly versatile family of ncRNA molecules.
Acknowledgments
We thank Jennifer Gebetsberger for comments on the manuscript.
Funding: We are grateful for the support of tRNA-derived fragment research in our laboratory by the NCCR ‘RNA & Disease’ funded by the Swiss National Science Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Guoping Li (School of Life Sciences and Technology, Tongji University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.08.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Borek E, Baliga BS, Gehrke CW, et al. High turnover rate of transfer RNA in tumor tissue. Cancer Res 1977;37:3362-6. [PubMed]
- Levitz R, Chapman D, Amitsur M, et al. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J 1990;9:1383-9. [PubMed]
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 2005;280:42744-9. [Crossref] [PubMed]
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008;14:2095-103. [Crossref] [PubMed]
- Yamasaki S, Ivanov P, Hu GF, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009;185:35-42. [Crossref] [PubMed]
- Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391-6. [Crossref] [PubMed]
- Gebetsberger J, Wyss L, Mleczko AM, et al. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 2016;1-10. [Crossref] [PubMed]
- Gebetsberger J, Zywicki M, Kunzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012;2012:260909.
- Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011;43:613-23. [Crossref] [PubMed]
- Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010;285:10959-68. [Crossref] [PubMed]
- Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 2014;34:2450-63. [Crossref] [PubMed]
- Goodarzi H, Liu X, Nguyen HC, et al. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015;161:790-802. [Crossref] [PubMed]
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A 2015;112:E3816-25. [Crossref] [PubMed]
- Venkatesh T, Suresh PS, Tsutsumi R. tRFs: miRNAs in disguise. Gene. 2016;579:133-8. [Crossref] [PubMed]
- Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397-400. [Crossref] [PubMed]
- Schorn AJ, Gutbrod MJ, LeBlanc C, et al. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017;170:61-71.e11. [Crossref] [PubMed]
- Hyun S. Small RNA Pathways That Protect the Somatic Genome. Int J Mol Sci 2017;18:E912 [Crossref] [PubMed]
- Yeung ML, Bennasser Y, Watashi K, et al. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009;37:6575-86. [Crossref] [PubMed]
- Ruggero K, Guffanti A, Corradin A, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol 2014;88:3612-22. [Crossref] [PubMed]
- Garcia-Silva MR, Frugier M, Tosar JP, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 2010;171:64-73. [Crossref] [PubMed]
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013;10:1798-806. [Crossref] [PubMed]
- Ogawa T. tRNA-targeting ribonucleases: molecular mechanisms and insights into their physiological roles. Biosci Biotechnol Biochem 2016;80:1037-45. [Crossref] [PubMed]
- Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, et al. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Reports 2017;8:112-24. [Crossref] [PubMed]
- Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014;33:2020-39. [Crossref] [PubMed]
- Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA 2017;8. [PubMed]
Cite this article as: Cristodero M, Polacek N. The multifaceted regulatory potential of tRNA-derived fragments. Non-coding RNA Investig 2017;1:7.


