Circular RNA in cardiovascular disease
Introduction
Over the past 20 years, different approaches have been developed to improve the diagnosis and treatment of cardiovascular disease (CVD) (1-3), among which translational medicine from basic research to clinical research plays an important role. However, they are still far from satisfactory. Thus, there is still a clinical need for investigating novel diagnostic biomarkers and new therapeutic targets to decrease the morbidity and mortality as well as improve the quality of life, considering the unsatisfactory outcome of the human disease.
Circular RNAs (circRNAs) are a recently discovered non-coding RNAs, which have been shown to be involved in multiple physiological and pathological processes including early embryonic development (4), aging (5) and disease-related pathways. Moreover, circRNAs can not only be found in different tissues (6,7), organs (8,9) but also enriched in body fluids (10-12), such as blood, exosomes, saliva, seminal plasma, indicating that circRNAs are promising therapy targets and promising biomarkers for human disease.
Specifically, circRNAs are widely expressed in the heart of human (9,13), rats (13) and mice (13,14) and have been shown to be associated with CVDs. In this review, we summarize the biogenesis and expression of circRNA, their function mechanism, the common software or online tool for circRNA research. Specially, we discuss circRNAs and their potential roles in diagnosis, prognosis and therapeutic strategies for CVDs.
Biogenesis and expression of circRNA (Figure 1)
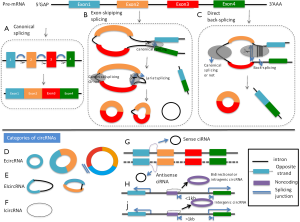
In unicellular organisms, circRNAs mostly stem from self-splicing introns of pre-ribosomal RNA (15,16), but can also arise from protein-coding genes in archaea (15). In the validated circRNAs in animals, circRNAs are predominantly derived from exons, whose 5’ and 3’ ends are covalently linked to form a “head-to-tail” splice junction. Although spliceosomal machinery is implicated in the pre-mRNA splicing (17), the mechanisms involved in exon circularization remain elusive.
It is supposed that circRNA generated from the editing process of pre-mRNA. There are two major mechanisms of circRNA formation: (I) direct back-splicing and (II) exon skipping (18,19). Direct back-splicing, where exons are spliced in non-canonical order, connects the downstream 5’ splice site (splice donor) to an upstream 3’ splice site (splice acceptor), thereby generating a circular transcript (20). The second mechanism is lariat splicing, which involves generation of a lariat intermediate containing exons, and then, the introns in the lariat are removed, generating exonic circRNA (ecircRNAs) (21). Both mechanisms function in vivo and correlate with the canonical spliceosome, but direct back-splicing may occur more frequently than exon skipping in ecircRNA biogenesis (22). Recent findings also indicate that RNA pairing, which can be formed by either repetitive elements (23) or non-repetitive but complementary sequences (15), across flanking introns is positively associated with exon circularization. Additional observations have revealed that RNA binding proteins such as adenosine to inosine acting on RNA enzyme (ADAR), muscleblind (MBL) and Quaking (QKI) function in specific ecircRNA biogenesis (24-26). Of note, these three proteins are known to be implicated in alternative splicing. In addition, compared with all expressed exons, the exons generating single-exon circRNA are 3-fold longer in length (6,23). Therefore, the length of a given exon may also correlate with circularization.
Based on the above progress of circRNA formation, circRNAs have 5 categories: (I) ecircRNAs, spliced from the exonic segments; (II) intronic circRNA (icircRNAs), derived wholly from an intron of the linear transcript; (III) exon-intron circRNA (EIicRNA), consisted of both exons and introns; (IV) antisense, contained one or more exons of the linear transcript on the opposite strand; (V) bidirectional or intragenic, transcribed from same gene locus of the linear transcript but in close genomic proximity and not classified as “sense” and “intronic”; and (VI) intergenic, when it is located in the genomic interval between two genes (Figure 1).
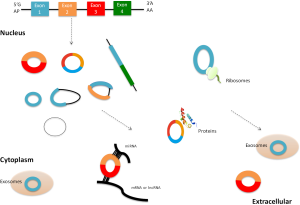
While most circRNAs are lowly abundant, some are ubiquitously expressed and are present at higher copy numbers (>10-fold) as compared to their linear transcripts (7,23). Besides, the levels of some circRNAs are significantly associated with the proliferative and differentiative stage (8,27,28), indicating that circRNAs may have important roles in organic development and functions. What’s more, circRNAs are highly stable in vivo compared with their linear counterparts, and are predominantly in the cytoplasm and can be sorted into exosomes (11). Considering their widely distribution in body fluid (10-12), circRNAs can sever as a promising biomarker for human disease, including CVDs.
Biogenetic function of circRNA: old tree with new flower
CircRNAs are attracting more attention owing to not only their abundance in the eukaryotic transcriptomes but also their functions. So far, it is regarded that circRNAs can function in multiple biological processes, such as miRNA binding, protein binding and regulation of transcription and post-transcription (Figure 2).
miRNA sponge
The most exciting finding is that circRNAs could function as microRNA (miRNA) sponges. For instance, ciRS-7/CDR1as harbors >70 conserved binding sites for miR-7 (29,30). miR-7 activity could be dramatically reduced through tethering of this miRNA to ciRS-7/CDR1as, resulting in increased levels of miR-7 targets. Then, circRNA destruction could release a shower of miR-7. Sry, a testis-specific circRNA, is another validated miRNA sponge (30,31). This circRNA harbors 16 binding sites for miR-138. These findings changed the mechanistic understanding of miRNA regulatory networks and increased the complexity of competitive endogenous RNA (ceRNA) network (32). Although few circRNAs contain a majority of miRNA binding sites for a single miRNA, the depletion of polymorphisms at predicted miRNA binding sites implies that circRNAs could be efficiently targeted by miRNAs.
Protein sponge
CircRNAs can also act as protein sponges. For example, circMbl, derived from the muscleblind (MBL/MBNL1) in flies and humans, harbors multiple muscleblind binding sites (25). When MBL proteins are in excess, circMbl could sponge out the redundant ones. By this way, MBL levels could be well regulated. And circ-Foxo3, which is downregulated in cancer cells and is associated with cell cycle progression, also interacts with some proteins. For examples, circ-Foxo3 binds cyclin-dependent kinase 2 (CDK2) and p21 (CDKN 1a), forming an RNA-protein complex that disrupts the interactions of CDK2 with cyclins A and E, required for cell cycle progression (33). Circ-Foxo3 also interacts with proteins inhibitors of differentiation (ID1), E2F transcription factor 1 (E2F1), focal adhesion kinase (FAK) and hypoxia inducible factor-1 (HIF-1), retaining them in the cytoplasm and thus promoting cardiac senescence (34).
Alternative splicing, mRNA trap and transcriptional regulation
CircRNAs can affect their parental genes in cis- or transactions. Circularization and splicing compete against each other, enabling ecircRNAs to function in alternative splicing (25). Once a back-splice occurs, it removes the internal exons, causing an alternative splicing. EcircRNAs may serve as an “mRNA trap” by sequestering the translation start site, making the truncated linear mRNA fail to translate. For instance, the mice formin (Fmn) gene could generate ecircRNAs acting as mRNA trap. Deletion of specific exons caused a failure to produce these ecircRNAs, but appeared to produce normal amounts of the linear mRNAs. The inability to generate ecircRNAs from the Fmn locus caused aberrant expression of the formin protein and changed the phenotype as a result (25).
Moreover, ciRNAs can function as a positive regulator of Pol II transcription by interacting with Pol II machinery (35). More recently, a study has revealed that EIciRNAs use a specific interaction with U1 snRNA to fine-tune the expression of their parental genes in the nucleus (36). This discovery not only reveals a role for circRNAs in transcriptional control but also highlights a regulatory strategy through specific RNA–RNA interaction between EIciRNA and U1 snRNA.
Collectively, the interplay between circRNAs and the transcriptional machinery provides new insights into the strategies to regulate gene expression in cells.
circRNAs and cardiovascular disease: common molecules with huge potentials (Table 1)
Table 1
| Disease | circRNAs | Model | Expression | Possible targets | Potential action | Potential value |
|---|---|---|---|---|---|---|
| MI (37) | Cdr1as (CiRS-7) | Mice | Increased | SP1, miR-7a; PARP | Overexpression of Cdr1as aggravated MI injuries with upregulated PARP and SP1, which could be rescued by the co-overexpression of miR-7a | Therapeutic, target |
| AMI (38) | Circ-081881 | Patients | Decreased | miR-548 | CircRNA_081881 could compete for miR-548 with PPARγ | Biomarker and Therapeutic |
| MI with or without left; ventricular dysfunction (39) | MIRC | Patients | Decreased | – | MICRA, can predict LV dysfunction after 3 to 4 months | Biomarker |
| CAD (40) | ANRIL | Patients | Increased | PES1 | circANRIL confers atheroprotection by controlling ribosomal RNA (rRNA) maturation and modulating pathways of atheroprotection | Therapeutic, target |
| Heart failure (41) | HRCR | Mice | Increased | miR-223, ARC | HRCR inhibits cardiac hypertrophy through miR-223 | Therapeutic, target |
| Cardiomyopathy (34) | circFOX-3 | Patients | Increased | ID-1, E2F1, FAK, HIF-1α | circ-Foxo3 induce senescence by interacting with the anti-senescent protein ID-1 and the transcription factor E2F1, as well as the anti-stress proteins FAK and HIF-1α | Therapeutic, target |
| Aortic aneurysm (42) | Circ-000595 | Patients | Increased | miR-19a | hsa-circ-000595 was highly expressed aortic tissues of patients with aortic aneurysm | Therapeutic, target |
| Hypoxic conditions (43) | cZNF-292 | Endothelial cell | – | Exhibits proangiogenic activities | Therapeutic, target |
MI, myocardial infarction; PARP, poly (ADPribose) polymerase; AMI, acute myocardial infarction; CAD, coronary artery disease; PES1, Pescadillo homologue 1; HRCR, heart-related circRNA; ARC, apoptosis repressor with caspase recruitment domain; ID1, inhibitors of differentiation; E2F1, E2F transcription factor 1; FAK, focal adhesion kinase; HIF-1, hypoxia inducible factor-1.
Since Nature firstly ascribed an actual function to one of these circRNA molecules (29), circRNAs become a research hotspot in the field of RNA. Though circRNAs are old molecules, the huge potential role of circRNAs are recently being discovered from the laboratory to the clinic. CircRNAs have been shown to be associated with various diseases in both patients’ cohorts and animal models including CVDs. Table 1 summarizes circRNAs that have been reported in CVDs.
CircRNAs-miRNAs axes in CVDs
As the miRNA sponge, several studies demonstrate the key role of circRNA’s in regulating miRNA in CVD.
Heart failure
A heart-related circRNA (HRCR) was reported to be able to attenuate the development of cardiac hypertrophy and heart failure in mice by targeting miR-223 and ARC (apoptosis repressor with caspase recruitment domain) (41). This study revealed a novel cardiac hypertrophic regulating model that was composed of HRCR, miR-223 and ARC. miR-223 promote the process of cardiac hypertrophy and heart failure, while ARC is the downstream of miR-223. However, the pathological effects of miR-223 can be inhibited by HRCR, which is identified as an upstream regulator of miR-223.
Myocardial infarction (MI)
In MI mice, Cdr1as (or CiRS-7) (37) was reported to be upregulated with increased cardiac infarct size. In Geng’s study, Cdr1as and miR-7 were both overexpressed in MI mice, but the function of Cdr1as can be downregulated by miR-7. SP1 and poly (ADP-ribose) polymerase (PARP) were the downstream targets of miR-7.
Aortic aneurysm
Aortic aneurysm is a leading cause of cardiovascular mortality. CircRNA has-circ-000595 was highly expressed in tissues from aortic aneurysm patients and hypoxic aortic smooth muscle cells (42). Knockdown of hsa-circ-000595 resulted in decreased apoptotic rate of human aortic smooth muscle cells. It was determined that miR-19a was a target of hsa-circ-000595.
Cardiac fibrosis
Individuals with diabetes are at significantly risk of developing diabetic myocardial fibrosis. CircRNA_000203 was up-regulated in the diabetic mouse myocardium and in Ang-II-induced mouse cardiac fibroblasts (44). CircRNA_000203 containing two potential binding sites for microRNA-26b-5p(miR-26b-5p), which exerted the anti-fibrotic effect by targeting Colla2 and CTGF. Over-expression of circRNA_000203 could eliminate the anti-fibrotic effect of miR-26b-5p in cardiac fibroblasts. Furthermore, blocking the function of miR-26b-5p contributed to the pro-fibrosis effect of circRNA_000203 in cardiac fibroblasts.
CircRNA-protein axes associated with CVDs
Coronary atherosclerosis
ANRIL (antisense non-coding RNA in the INK4 locus) was differentially expressed by the genotype at 9p21 (45) and increased linear ANRIL (linANRIL) was associated with increased atherosclerosis (46). ANRIL can also be expressed as circular transcripts, such as circANRIL. CircANRIL can protect the tissue from atherosclerosis by controlling ribosomal RNA (rRNA) maturation and modulation pathways of atherogenesis (40). Pescadillo homologue 1 (PES1), which was an essential 60S-preribosomal assembly factor, was the downstream target of circANRIL. CircANRIL can impair exonuclease-mediated pre-rRNA processing and ribosome biogenesis in vascular smooth muscle cells and macrophages through binding to PES1. Overall, these findings demonstrated that circANRIL plays an important role in regulating ribosome biogenesis and conferring atheroprotection.
Cardiomyopathy
circ-Foxo3 was highly expressed in heart samples of aged patients and mice, and was shown to be correlated with cellular senescence (34). Doxorubicin-induced cardiomyopathy was aggravated by ectopic expression of circ-Foxo3 but was relieved by silencing endogenous circ-Foxo3. circ-Foxo3 induce senescence by interacting with the anti-senescent protein ID-1 and the transcription factor E2F1, as well as the anti-stress proteins FAK and HIF-1α. Strikingly, in their study, in vivo delivery of circ-Foxo3 targeted siRNA into mouse embryonic fibroblasts and primary cardiac myocytes resulted in lower levels of circ-Foxo3 expression and beta-gal staining, which is a standard marker for senescence, suggesting a potential therapeutic approach for myocardial protection.
Except for those circRNAs that have been shown the interaction with miRNAs or proteins, there’re also some circRNAs correlated with CVDs, which have not been found the functional mechanisms (14,43). cZNF292 is a hypoxic-induced circRNA, which has been shown to be regulated hypoxia and have biological functions (43), however, no validated microRNA-binding sites for cZNF292 were detected, suggesting that cZNF292 does not act as a microRNA sponge. The circRNA landscape of heart tissue can be explored using a novel DCC software (14). Among those circRNAs, several circRNAs can be directly attributed to host genes that have been previously described as associated with CVD. Further studies of these candidate circRNAs are needed to reveal disease-relevant properties or functions of specific circRNAs.
CircRNA as biomarker for CVD
In addition to participate to the pathophysiological processes of CVD, circRNA has also been shown to be biomarkers for human disease, including circRNAs from pathological section (47,48), blood plasma (49,50), saliva (51) or seminal fluid (12). Several studies have been taken out to find out whether circulating circRNAs can be biomarkers for CVD (38,39).
For example, circRNA have the potential to predict outcome after reperfusion (39). In this study, they enrolled patients diagnosed as MI with or without left ventricular dysfunction, and finally identified a circRNA, MICRA, can predict LV dysfunction after 3 to 4 months. CircRNA can be also used as a potential target for the precision diagnosis and therapy of AMI (38). This study demonstrated that circRNA_081881 regulated PPARγ expression by functioning as a competing endogenous RNA (ceRNA) of miR-548. Considering that circRNA_081881 was detected in the plasma samples, circRNA_081881 may be used as a potential target for AMI.
In addition, some other circRNAs have been shown to be associated with CAD. For instance, that the expression of hsa_circ_0124644 was moderately correlated with the CAD (52) implying that hsa_circ_0124644 might be deeply involved in the pathologies of CAD. As hsa_circ_0124644 can be easily determined using peripheral blood at a relatively low cost, yet the specificity and sensitivity are relatively high, and the diagnostic value would slightly increase after introducing hsa_circ_0098964, making hsa_circ_0124644 a powerful tool in the diagnosis of CAD.
Characters of circRNAs as biomarker for CVD
CircRNAs are widely found in blood, saliva, and seminal fluid, and they are stable due to their circular structure, which are pre-conditions for their usage as biomarkers for various diseases. Furthermore, the expression levels of circRNAs vary under different conditions, making them possible to be biomarkers for diseases.
At present, molecular biomarkers used in the clinic are always proteins that have low organ specificity. For example, atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP) are common in heart failure, and cardiac troponin T (cTnT) is common in MI. If circRNAs can be used in the clinic as biomarkers of CVD, their specific expression may assist in solving the problem of existing markers’ low organ specificity.
However, there’re still some questions remains to be solved. First, the expression of some circRNAs only differs in patients’ tissue, but not in patients’ blood, which may cause some trauma for patients. Second, the relationship between the expression levels of circRNAs and the severity of CVD remains unknown, and thus we cannot estimate the severity of disease according to the expression levels of circRNAs. Third, the reliability of using circRNAs for diagnosis still needs to be proven. Furthermore, due to the enormous number of circRNAs, their functions may be complex and different from each other. Current studies mainly focus on the interactions between RNAs and proteins, but the function of their secondary structures and their roles in DNA genome are still unknown.
Conclusions
Considering the facts that circRNA can act as miRNA or protein sponges and are widely expressed in the heart of human, mice and rats, it is plausible that some circRNAs can act as local regulators of gene expression. Although some circRNAs have been proved to be the regulators of pathophysiological processes in CVD, much more functional circRNA studies are needed to dissect their roles and mechanism-of-action in the CVD and development of cardiovascular biology.
In addition to a void of data on the functional roles of circRNAs, circRNAs also have the potential to be a biomarker for CVD. The specific expression of circRNAs may assist in solving the problem of existing markers’ low organ specificity. Although not discussed in detail in the current review, circRNAs are also viewed as targets for designing therapeutic approaches in CVD. For example, circANRIL induced apoptosis and inhibited proliferation by binding to PES1△446-588 (40), therefore, it is plausible that a drug can be produced to competitive bind PES1△446-588 and thus escape such a negative effect.
In conclusion, circRNAs may play an important role in CVD, and are potential biomarkers or therapy targets for CVDs.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (NO. 812201080003).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.08.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nioi P, Sigurdsson A, Thorleifsson G, et al. Variant ASGR1 Associated with a Reduced Risk of Coronary Artery Disease. N Engl J Med 2016;374:2131-41. [Crossref] [PubMed]
- Abraham G, Havulinna AS, Bhalala OG, et al. Genomic prediction of coronary heart disease. Eur Heart J 2016;37:3267-78. [Crossref] [PubMed]
- Yusuf S, Bosch J, Dagenais G, et al. Cholesterol Lowering in Intermediate-Risk Persons without Cardiovascular Disease. N Engl J Med 2016;374:2021-31. [Crossref] [PubMed]
- Fan X, Zhang X, Wu X, et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 2015;16:148. [Crossref] [PubMed]
- Wang YH, Yu XH, Luo SS, et al. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing 2015;12:17. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [PubMed]
- Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015;61:221-30. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Dong WW, Li HM, Qing XR, et al. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci Rep 2016;6:39080. [Crossref] [PubMed]
- Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. [Crossref] [PubMed]
- Jakobi T, Czaja-Hasse LF, Reinhardt R, et al. Profiling and Validation of the Circular RNA Repertoire in Adult Murine Hearts. Genomics Proteomics Bioinformatics 2016;14:216-23. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009;136:701-18. [Crossref] [PubMed]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015;4:e07540 [Crossref] [PubMed]
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015;21:172-9. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [PubMed]
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93:6536-41. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Venø MT, Hansen TB, Veno ST, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015;16:245. [Crossref] [PubMed]
- You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015;18:603-10. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993;73:1019-30. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017;38:1402-12. [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753 [Crossref] [PubMed]
- Deng YY, Zhang W, She J, et al. Circular RNA Related to PPARg Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J Am Coll Cardiol 2016;68:C51-C2. [Crossref]
- Vausort M, Salgado-Somoza A, Zhang L, et al. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J Am Coll Cardiol 2016;68:1247-8. [Crossref] [PubMed]
- Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429. [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Zheng C, Niu H, Li M, et al. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep 2015;12:6656-62. [Crossref] [PubMed]
- Boeckel JN, Jae N, Heumuller AW, et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res 2015;117:884-90. [Crossref] [PubMed]
- Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342. [Crossref] [PubMed]
- Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol 2012;32:196-206. [Crossref] [PubMed]
- Holdt LM, Beutner F, Scholz M, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol 2010;30:620-7. [Crossref] [PubMed]
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015;6:6001-13. [Crossref] [PubMed]
- Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016;16:161-9. [Crossref] [PubMed]
- Zhao Z, Li X, Jian D, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol 2017;54:237-45. [Crossref] [PubMed]
- Cui X, Niu W, Kong L, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med 2016;10:943-52. [Crossref] [PubMed]
- Lin X, Lo HC, Wong DT, et al. Noncoding RNAs in human saliva as potential disease biomarkers. Front Genet 2015;6:175. [Crossref] [PubMed]
- Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918. [Crossref] [PubMed]
Cite this article as: Wang X, Ding Z, Wu J, Wang S, Zou Y. Circular RNA in cardiovascular disease. Non-coding RNA Investig 2017;1:5.