
CircCCDC66: the colorectal oncogene
Circular RNAs (circRNAs) are a large class of long non-coding RNAs produced by backsplicing or “head-to-tail” splicing (1). Due to the lack of free ends, circRNAs escape the conventional exonucleolytic RNA degradation pathways and thus have much higher stability than linear RNAs. The high stability allows the circRNAs to accumulate to high levels and conduct functions for prolonged periods in cells (2-4). Specific circRNAs have been functionally characterized as protein sponges, miRNA sponges, protein scaffolds or regulators of splicing and transcription (5-7). Many circRNAs are differentially expressed in cancerous tissues correlating with cancer progression (8,9). Among these, the circRNAs circFOXO3 and circITHC have been identified as tumor-suppressor circRNAs that are downregulated in tumor tissues. For both circRNAs, it has been suggested that they function as miRNA sponges to protect tumor-suppressor genes from miRNA attack (10,11) (Figure 1). However, the function of most circRNAs remains elusive although many of them have a high potential as biomarkers in cancer.
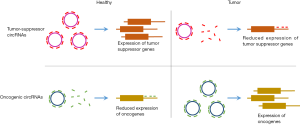
Recently, Hsiao et al. published a study revealing a novel oncogenic function of the circRNA circCCDC66 in colorectal cancer progression (12). They developed a pipeline to identify highly expressed circRNAs in RNA sequencing data, and from this, the authors found circCCDC66 as a novel cancer associated circRNA highly upregulated in colorectal cancer. Specifically, they found that 88% of the colorectal cancer patients had elevated levels of circCCDC66 and that the overall survival rate was lower for patients with high circCCDC66 compared to patients with low expression. Hence, circCCDC66 has promising prognostic biomarker potential. The elevated expression of circCCDC66 in colorectal cancer indicates an important function in tumor development. To validate this hypothesis, Hsiao et al. performed knockdown and overexpression of circCCDC66. Knockdown of circCCDC66 in tumor cell cultures was conducted with siRNAs targeting the backsplice junction specifically. This resulted in a decreased tumor cell proliferation, migration and invasion. Furthermore, using xenograft mouse models, depletion of circCCDC66 prior to transplantation showed reduced tumor aggression. These observations were supported by increased tumor cell proliferation, colony formation and DNA synthesis upon circCCDC66 overexpression, strongly implicating circCCDC66 as an important driver in cancer etiology. Consistent with previous studies of circRNAs acting as miRNA sponges in normal and cancerous tissues, the authors performed a miRNA target sequence analysis. They found that circCCDC66 has several target sites for different tumor-suppressor miRNAs. Hence, overexpression of circCCDC66 increased the expression of oncogenes by tethering and inhibiting the activity of miRNAs otherwise involved in oncogene repression. In addition, knockdown of circCCDC66 resulted in decreased expression of oncogenes targeted by miRNAs released from circCCDC66-mediated sponging. As a remarkable good control, simultaneous anti-miR inhibition of the involved miRNAs rescued the effect of circRNA knockdown. Overall, the experiments led to the conclusion that circCCDC66 functions as an oncogenic circRNA by sponging several important tumor-suppressor miRNAs.
The impressive work by Hsiao et al. provides important knowledge and insights into the field of circRNAs and identifies a promising biomarker and potential therapeutic target, circCCDC66, in colorectal cancer. While the evidence of circRNAs acting as miRNA sponges is growing almost on a daily basis, it clashes with the stoichiometric modelling of circRNA: miRNA: mRNA levels required for efficient sponge effects (13-15) and the abundancy and conservation of miRNA target sites found in circRNAs in general (2). Clearly, there are aspects of the complex regulatory networks within cells that are well outside our current grasp and knowledge, and hopefully in the years to come, more eminent RNA research will settle the controversy and establish a novel paradigm in which the bioinformatics modelling and the experimental data are aligned with consistent output.
Acknowledgments
Funding: This work was supported by the Novo Nordisk Foundation (NNF16OC0019874).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dongchao Lu [IFB, Molecular and Translational Therapeutic Strategies (Imtts), Hannover Medical School, Hanover, Germany].
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.08.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8.
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103-11. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Molecular cell 2014;56:55-66. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015;16:4. [Crossref] [PubMed]
- Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 2015;5:8057. [Crossref] [PubMed]
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001-13. [Crossref] [PubMed]
- Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2016;35:3919-31. [Crossref] [PubMed]
- Hsiao KY, Lin YC, Gupta SK, et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res 2017;77:2339-50. [Crossref] [PubMed]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. [Crossref] [PubMed]
- Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 2015;16:113-26. [Crossref] [PubMed]
- Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell 2014;56:347-59. [Crossref] [PubMed]
Cite this article as: Nielsen KM, Hansen TB. CircCCDC66: the colorectal oncogene. Non-coding RNA Investig 2017;1:3.


